6B9P
 
 | | Structure of GH 38 Jack Bean alpha-mannosidase in complex with a 36-valent iminosugar cluster inhibitor | | Descriptor: | (2R,3R,4R,5S)-2-(hydroxymethyl)-1-{9-[4-(methoxymethyl)-1H-1,2,3-triazol-1-yl]nonyl}piperidine-3,4,5-triol, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-mannosidase from Canavalia ensiformis (jack bean), ... | | Authors: | Howard, E, Cousido-Siah, A, Lepage, M, Bodlenner, A, Mitschler, A, Meli, A, De Riccardis, F, Izzo, I, Podjarny, A, Compain, P. | | Deposit date: | 2017-10-11 | | Release date: | 2018-09-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | Structural Basis of Outstanding Multivalent Effects in Jack Bean alpha-Mannosidase Inhibition.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
2AGT
 
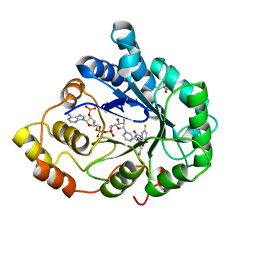 | | Aldose Reductase Mutant Leu 300 Pro complexed with Fidarestat | | Descriptor: | (2S,4S)-2-AMINOFORMYL-6-FLUORO-SPIRO[CHROMAN-4,4'-IMIDAZOLIDINE]-2',5'-DIONE, Aldose reductase, CHLORIDE ION, ... | | Authors: | Petrova, T, Steuber, H, Hazemann, I, Cousido-Siah, A, Mitschler, A, Chung, R, Oka, M, Klebe, G, El-Kabbani, O, Joachimiak, A, Podjarny, A. | | Deposit date: | 2005-07-27 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Factorizing Selectivity Determinants of Inhibitor Binding toward Aldose and Aldehyde Reductases: Structural and Thermodynamic Properties of the Aldose Reductase Mutant Leu300Pro-Fidarestat Complex
J.Med.Chem., 48, 2005
|
|
6Q39
 
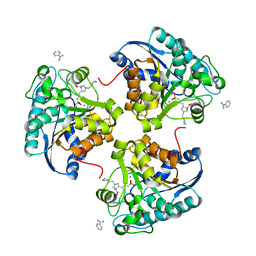 | | Complex of Arginase 2 with Example 49 | | Descriptor: | 3-[(3~{S},4~{R})-4-azanyl-4-carboxy-1-[[(2~{S})-piperidin-2-yl]methyl]pyrrolidin-3-yl]propyl-tris(oxidanyl)boranuide, Arginase-2, mitochondrial, ... | | Authors: | Podjarny, A.D, Van Zandt, M.C, Cousido-Siah, A. | | Deposit date: | 2018-12-03 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.213 Å) | | Cite: | Discovery ofN-Substituted 3-Amino-4-(3-boronopropyl)pyrrolidine-3-carboxylic Acids as Highly Potent Third-Generation Inhibitors of Human Arginase I and II.
J.Med.Chem., 62, 2019
|
|
6Q37
 
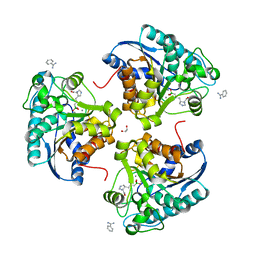 | | Complex of Arginase 2 with Example 23 | | Descriptor: | 1,2-ETHANEDIOL, 3-[(3~{S},4~{R})-4-azanyl-4-carboxy-pyrrolidin-3-yl]propyl-tris(oxidanyl)boranuide, Arginase-2, ... | | Authors: | Podjarny, A.D, Van Zandt, M.C, Cousido-Siah, A. | | Deposit date: | 2018-12-03 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.211 Å) | | Cite: | Discovery ofN-Substituted 3-Amino-4-(3-boronopropyl)pyrrolidine-3-carboxylic Acids as Highly Potent Third-Generation Inhibitors of Human Arginase I and II.
J.Med.Chem., 62, 2019
|
|
5CE5
 
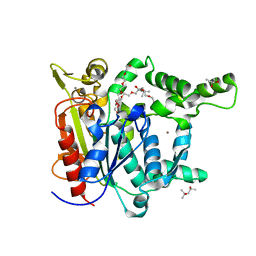 | | Probing the roles of two tryptophans surrounding the unique zinc coordination site in lipase family I.5 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, CALCIUM ION, ... | | Authors: | Sezerman, O.U, Podjarny, A.D, Emel, T, Cousido-Siah, A, Mitschler, A. | | Deposit date: | 2015-07-06 | | Release date: | 2015-12-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the roles of two tryptophans surrounding the unique zinc coordination site in lipase family I.5.
Proteins, 84, 2016
|
|
7R2T
 
 | | SYNJ2BP complex with a phosphorylated Vangl2 peptide at the P-1 position. | | Descriptor: | CALCIUM ION, GLYCEROL, Synaptojanin-2-binding protein,Annexin, ... | | Authors: | Carrasco, K, Cousido Siah, A, Gogl, G, Betzi, S, McEwen, A, Kostmann, C, Trave, G. | | Deposit date: | 2022-02-05 | | Release date: | 2023-05-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | SYNJ2BP PDZ domain in complex with a synthetic Vangl2 peptide.
To Be Published
|
|
7P73
 
 | | The PDZ domain of SYNJ2BP complexed with the PDZ-binding motif of HTLV1-TAX1 | | Descriptor: | CALCIUM ION, GLYCEROL, Protein Tax-1, ... | | Authors: | Gogl, G, Cousido-Siah, A, Trave, G. | | Deposit date: | 2021-07-19 | | Release date: | 2022-07-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Quantitative fragmentomics allow affinity mapping of interactomes.
Nat Commun, 13, 2022
|
|
7P74
 
 | | The PDZ domain of SYNJ2BP complexed with the phosphorylated PDZ-binding motif of RSK1 | | Descriptor: | CALCIUM ION, GLYCEROL, Ribosomal protein S6 kinase alpha-1, ... | | Authors: | Gogl, G, Cousido-Siah, A, Trave, G. | | Deposit date: | 2021-07-19 | | Release date: | 2022-07-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Quantitative fragmentomics allow affinity mapping of interactomes.
Nat Commun, 13, 2022
|
|
7P70
 
 | | The PDZ-domain of SNTB1 complexed with the PDZ-binding motif of HPV35-E6 | | Descriptor: | Beta-1-syntrophin,Annexin A2, CALCIUM ION, GLYCEROL, ... | | Authors: | Gogl, G, Cousido-Siah, A, Trave, G. | | Deposit date: | 2021-07-19 | | Release date: | 2022-07-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Quantitative fragmentomics allow affinity mapping of interactomes.
Nat Commun, 13, 2022
|
|
7P71
 
 | | The PDZ domain of MAGI1_2 complexed with the PDZ-binding motif of HPV35-E6 | | Descriptor: | CALCIUM ION, CITRIC ACID, GLYCEROL, ... | | Authors: | Gogl, G, Cousido-Siah, A, Trave, G. | | Deposit date: | 2021-07-19 | | Release date: | 2022-07-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Quantitative fragmentomics allow affinity mapping of interactomes.
Nat Commun, 13, 2022
|
|
7P72
 
 | | The PDZ domain of SNX27 complexed with the PDZ-binding motif of MERS-E | | Descriptor: | CALCIUM ION, Envelope small membrane protein, GLYCEROL, ... | | Authors: | Gogl, G, Cousido-Siah, A, Trave, G. | | Deposit date: | 2021-07-19 | | Release date: | 2022-07-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Quantitative fragmentomics allow affinity mapping of interactomes.
Nat Commun, 13, 2022
|
|
8AEL
 
 | | SYNJ2BP complex with a synthetic Vangl2 peptide (3mer). | | Descriptor: | CALCIUM ION, GLY-GLY-GLY-THR-SER-VAL, GLYCEROL, ... | | Authors: | Carrasco, K, Cousido Siah, A, Gogl, G, Betzi, S, McEwen, A, Kostmann, C, Trave, G. | | Deposit date: | 2022-07-13 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | SYNJ2BP PDZ domain in complex with a synthetic Vangl2 peptide.
To Be Published
|
|
3QF6
 
 | | Neutron structure of type-III Antifreeze Protein allows the reconstruction of AFP-ice interface | | Descriptor: | Type-3 ice-structuring protein HPLC 12 | | Authors: | Howard, E.I, Blakeley, M.P, Haertlein, M, Petit-Haertlein, I, Mitschler, A, Fisher, S.J, Cousido-Siah, A, Salvay, A.G, Popov, A, Muller-Dieckmann, C, Petrova, T, Podjarny, A. | | Deposit date: | 2011-01-21 | | Release date: | 2011-06-22 | | Last modified: | 2024-03-20 | | Method: | NEUTRON DIFFRACTION (1.85 Å) | | Cite: | Neutron structure of type-III antifreeze protein allows the reconstruction of AFP-ice interface.
J.Mol.Recognit., 24, 2011
|
|
5NRF
 
 | | Crystal structure of human chitotriosidase-1 (hCHIT) catalytic domain in complex with compound 7i | | Descriptor: | 1-(3-azanyl-1~{H}-1,2,4-triazol-5-yl)-~{N}-[2-(4-chlorophenyl)ethyl]-~{N}-(phenylmethyl)piperidin-4-amine, Chitotriosidase-1, GLYCEROL | | Authors: | Mazur, M, Olczak, J, Olejniczak, S, Koralewski, R, Czestkowski, W, Jedrzejczak, A, Golab, J, Dzwonek, K, Dymek, B, Sklepkiewicz, P, Zagozdzon, A, Noonan, T, Mahboubi, K, Conway, B, Sheeler, R, Beckett, P, Hungerford, W.M, Podjarny, A, Mitschler, A, Cousido-Siah, A, Fadel, F, Golebiowski, A. | | Deposit date: | 2017-04-22 | | Release date: | 2018-03-28 | | Method: | X-RAY DIFFRACTION (1.447 Å) | | Cite: | Targeting Acidic Mammalian chitinase Is Effective in Animal Model of Asthma.
J. Med. Chem., 61, 2018
|
|
5EUC
 
 | | The role of the C-terminal region on the oligomeric state and enzymatic activity of Trypanosoma cruzi hypoxanthine phosphoribosyl transferase | | Descriptor: | Hypoxanthine-guanine phosphoribosyltransferase | | Authors: | Valsecchi, W.M, Cousido-Siah, A, Mitschler, A, Podjarny, A, Delfino, J.M, Santos, J. | | Deposit date: | 2015-11-18 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The role of the C-terminal region on the oligomeric state and enzymatic activity of Trypanosoma cruzi hypoxanthine phosphoribosyl transferase.
Biochim.Biophys.Acta, 1864, 2016
|
|
5HR3
 
 | | Crystal structure of thioredoxin N106A mutant | | Descriptor: | COPPER (II) ION, ETHANOL, SULFATE ION, ... | | Authors: | Noguera, M.E, Vazquez, D.S, Howard, E.I, Cousido-Siah, A, Mitschler, A, Podjarny, A, Santos, J. | | Deposit date: | 2016-01-22 | | Release date: | 2017-02-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.101 Å) | | Cite: | Structural variability of E. coli thioredoxin captured in the crystal structures of single-point mutants.
Sci Rep, 7, 2017
|
|
5HBF
 
 | | Crystal structure of human full-length chitotriosidase (CHIT1) | | Descriptor: | Chitotriosidase-1, GLYCEROL | | Authors: | Fadel, F, Zhao, Y, Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Podjarny, A. | | Deposit date: | 2015-12-31 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | X-Ray Crystal Structure of the Full Length Human Chitotriosidase (CHIT1) Reveals Features of Its Chitin Binding Domain.
Plos One, 11, 2016
|
|
5HR0
 
 | | Crystal structure of thioredoxin E101G mutant | | Descriptor: | COPPER (II) ION, Thioredoxin | | Authors: | Noguera, M.E, Vazquez, D.S, Howard, E.I, Cousido-Siah, A, Mitschler, A, Podjarny, A, Santos, J. | | Deposit date: | 2016-01-22 | | Release date: | 2017-02-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Structural variability of E. coli thioredoxin captured in the crystal structures of single-point mutants.
Sci Rep, 7, 2017
|
|
5HR1
 
 | | Crystal structure of thioredoxin L107A mutant | | Descriptor: | COPPER (II) ION, Thioredoxin-1 | | Authors: | Noguera, M.E, Vazquez, D.S, Howard, E.I, Cousido-Siah, A, Mitschler, A, Podjarny, A, Santos, J. | | Deposit date: | 2016-01-22 | | Release date: | 2017-02-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.144 Å) | | Cite: | Structural variability of E. coli thioredoxin captured in the crystal structures of single-point mutants.
Sci Rep, 7, 2017
|
|
2QXW
 
 | | Perdeuterated alr2 in complex with idd594 | | Descriptor: | Aldose reductase, CITRIC ACID, IDD594, ... | | Authors: | Blakeley, M.P, Ruiz, F, Cachau, R, Hazemann, I, Meilleur, F, Mitschler, A, Ginell, S, Afonine, P, Ventura, O, Cousido-Siah, A, Joachimiak, A, Myles, D, Podjarny, A. | | Deposit date: | 2007-08-13 | | Release date: | 2008-01-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | Quantum model of catalysis based on a mobile proton revealed by subatomic x-ray and neutron diffraction studies of h-aldose reductase.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2R24
 
 | | Human Aldose Reductase structure | | Descriptor: | Aldose reductase, IDD594, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Blakeley, M.P, Ruiz, F, Cachau, R, Hazemann, I, Meilleur, F, Mitschler, A, Ginell, S, Afonine, P, Ventura, O.N, Cousido-Siah, A, Haertlein, M, Joachimiak, A, Myles, D, Podjarny, A. | | Deposit date: | 2007-08-24 | | Release date: | 2008-12-23 | | Last modified: | 2024-02-21 | | Method: | NEUTRON DIFFRACTION (1.752 Å), X-RAY DIFFRACTION | | Cite: | Quantum model of catalysis based on mobile proton revealed by subatomic X-Ray and neutron diffraction studies of h-Aldose Reductase
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
6TWZ
 
 | | 14-3-3 sigma complexed with a phosphorylated 16E6 peptide | | Descriptor: | 14-3-3 protein sigma, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, D(-)-TARTARIC ACID, ... | | Authors: | Gogl, G, Cousido-Siah, A, Sluchanko, N.N, Trave, G. | | Deposit date: | 2020-01-13 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Dual Specificity PDZ- and 14-3-3-Binding Motifs: A Structural and Interactomics Study.
Structure, 28, 2020
|
|
6TWY
 
 | | MAGI1_2 complexed with a phosphomimetic RSK1 peptide | | Descriptor: | CALCIUM ION, CITRIC ACID, GLYCEROL, ... | | Authors: | Gogl, G, Cousido-Siah, A, Trave, G. | | Deposit date: | 2020-01-13 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Dual Specificity PDZ- and 14-3-3-Binding Motifs: A Structural and Interactomics Study.
Structure, 28, 2020
|
|
6TWU
 
 | | MAGI1_2 complexed with a phosphomimetic 16E6 peptide | | Descriptor: | CALCIUM ION, CITRIC ACID, GLYCEROL, ... | | Authors: | Gogl, G, Cousido-Siah, A, Trave, G. | | Deposit date: | 2020-01-13 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Dual Specificity PDZ- and 14-3-3-Binding Motifs: A Structural and Interactomics Study.
Structure, 28, 2020
|
|
6TWQ
 
 | | MAGI1_2 complexed with a 16E6 peptide | | Descriptor: | CALCIUM ION, CITRIC ACID, GLYCEROL, ... | | Authors: | Gogl, G, Cousido-Siah, A, Trave, G. | | Deposit date: | 2020-01-13 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Dual Specificity PDZ- and 14-3-3-Binding Motifs: A Structural and Interactomics Study.
Structure, 28, 2020
|
|
