4X66
 
 | | Crystal Structure of 30S ribosomal subunit from Thermus thermophilus | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Demirci, H, Chen, J, Choi, J, Soltis, M, Puglisi, J.D. | | Deposit date: | 2014-12-06 | | Release date: | 2015-11-18 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (3.446 Å) | | Cite: | N(6)-methyladenosine in mRNA disrupts tRNA selection and translation-elongation dynamics.
Nat.Struct.Mol.Biol., 23, 2016
|
|
4X64
 
 | | Crystal Structure of 30S ribosomal subunit from Thermus thermophilus | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Demirci, H, Chen, J, Choi, J, Soltis, M, Puglisi, J.D. | | Deposit date: | 2014-12-06 | | Release date: | 2015-11-18 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | N(6)-methyladenosine in mRNA disrupts tRNA selection and translation-elongation dynamics.
Nat.Struct.Mol.Biol., 23, 2016
|
|
4X62
 
 | | Crystal Structure of 30S ribosomal subunit from Thermus thermophilus | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Demirci, H, Chen, J, Choi, J, Soltis, M, Puglisi, J.D. | | Deposit date: | 2014-12-06 | | Release date: | 2015-11-18 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (3.4492 Å) | | Cite: | N(6)-methyladenosine in mRNA disrupts tRNA selection and translation-elongation dynamics.
Nat.Struct.Mol.Biol., 23, 2016
|
|
4X65
 
 | | Crystal Structure of 30S ribosomal subunit from Thermus thermophilus | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Demirci, H, Chen, J, Choi, J, Soltis, M, Puglisi, J.D. | | Deposit date: | 2014-12-06 | | Release date: | 2015-11-18 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (3.345 Å) | | Cite: | N(6)-methyladenosine in mRNA disrupts tRNA selection and translation-elongation dynamics.
Nat.Struct.Mol.Biol., 23, 2016
|
|
4YJY
 
 | |
3TVZ
 
 | | Structure of Bacillus subtilis HmoB | | Descriptor: | Putative uncharacterized protein yhgC | | Authors: | Choe, J, Choi, S, Park, S. | | Deposit date: | 2011-09-21 | | Release date: | 2012-07-11 | | Last modified: | 2019-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bacillus subtilis HmoB is a heme oxygenase with a novel structure.
Bmb Rep, 45, 2012
|
|
5WTS
 
 | |
4S11
 
 | |
4S10
 
 | |
1Z6X
 
 | | Structure Of Human ADP-Ribosylation Factor 4 | | Descriptor: | ADP-ribosylation factor 4, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Choe, J, Atanassova, A, Arrowsmith, C, Edwards, A, Sundstrom, M, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-03-23 | | Release date: | 2005-04-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure Of Human ADP-Ribosylation Factor 4
To be Published
|
|
1ZD9
 
 | | Structure of human ADP-ribosylation factor-like 10B | | Descriptor: | ADP-ribosylation factor-like 10B, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Choe, J, Atanassova, A, Arrowsmith, C, Edwards, A, Sundstrom, M, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-04-14 | | Release date: | 2005-05-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of human ADP-ribosylation factor-like 10B
to be published
|
|
1YZG
 
 | | Structure of Human ADP-ribosylation factor-like 8 | | Descriptor: | ADP-ribosylation factor-like 8, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Choe, J, Atanassova, A, Arrowsmith, C, Edwards, A, Sundstrom, M, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-02-28 | | Release date: | 2005-03-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Human ADP-ribosylation factor-like 8
To be Published, 2005
|
|
1Z6Y
 
 | | Structure Of Human ADP-Ribosylation Factor-Like 5 | | Descriptor: | ADP-ribosylation factor-like protein 5, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Choe, J, Atanassova, A, Arrowsmith, C, Edwards, A, Sundstrom, M, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-03-23 | | Release date: | 2005-04-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure Of Human ADP-Ribosylation Factor-Like 5
To be Published
|
|
1ZS6
 
 | | structure of human nucleoside-diphosphate kinase 3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Nucleoside diphosphate kinase 3 | | Authors: | Choe, J, Dimov, S, Arrowsmith, C, Edwards, A, Sundstrom, M, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-05-23 | | Release date: | 2005-06-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of human nucleoside-diphosphate kinase 3
To be Published
|
|
1ZJH
 
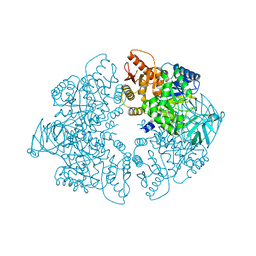 | | Structure of human muscle pyruvate kinase (PKM2) | | Descriptor: | Pyruvate kinase, isozymes M1/M2 | | Authors: | Choe, J, Atanassova, A, Arrowsmith, C, Edwards, A, Sundstrom, M, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-04-28 | | Release date: | 2005-05-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of human muscle pyruvate kinase (PKM2).
TO BE PUBLISHED
|
|
1ZIW
 
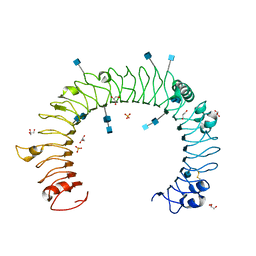 | | Human Toll-like Receptor 3 extracellular domain structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Wilson, I.A, Choe, J. | | Deposit date: | 2005-04-27 | | Release date: | 2005-06-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human toll-like receptor 3 (TLR3) ectodomain.
Science, 309, 2005
|
|
1ZD8
 
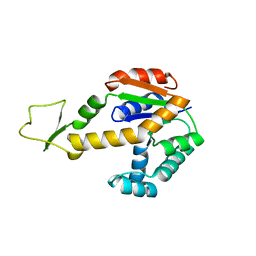 | | Structure of human adenylate kinase 3 like 1 | | Descriptor: | GTP:AMP phosphotransferase mitochondrial | | Authors: | Choe, J, Atanassova, A, Arrowsmith, C, Edwards, A, Sundstrom, M, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-04-14 | | Release date: | 2005-05-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structure of human adenylate kinase 3 like 1
to be published
|
|
6IM8
 
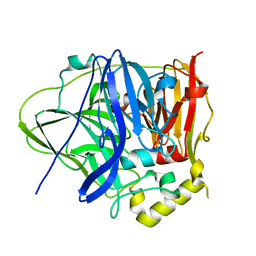 | | CueO-PM2 multicopper oxidase | | Descriptor: | Blue copper oxidase CueO,PM2 peptide,Blue copper oxidase CueO | | Authors: | Wongsantichon, J, Robinson, R, Ghadessy, F. | | Deposit date: | 2018-10-22 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Development and structural characterization of an engineered multi-copper oxidase reporter of protein-protein interactions.
J.Biol.Chem., 294, 2019
|
|
6IM9
 
 | | MDM2 bound CueO-PM2 sensor | | Descriptor: | Blue copper oxidase CueO,PM2 peptide,Blue copper oxidase CueO, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Wongsantichon, J, Robinson, R, Ghadessy, F. | | Deposit date: | 2018-10-22 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Development and structural characterization of an engineered multi-copper oxidase reporter of protein-protein interactions.
J.Biol.Chem., 294, 2019
|
|
6IM7
 
 | | CueO-12.1 multicopper oxidase | | Descriptor: | Blue copper oxidase CueO,12.1 peptide,Blue copper oxidase CueO, CALCIUM ION | | Authors: | Wongsantichon, J, Robinson, R, Ghadessy, F. | | Deposit date: | 2018-10-22 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Development and structural characterization of an engineered multi-copper oxidase reporter of protein-protein interactions.
J.Biol.Chem., 294, 2019
|
|
3OYY
 
 | |
4GCV
 
 | |
6WII
 
 | | Crystal structure of the K. pneumoniae LpxH/JH-LPH-41 complex | | Descriptor: | 5-({[4-({4-[3-chloro-5-(trifluoromethyl)phenyl]piperazin-1-yl}sulfonyl)phenyl]carbamoyl}amino)-N-hydroxypentanamide, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Cochrane, C.S, Cho, J, Fenton, B.A, Zhou, P. | | Deposit date: | 2020-04-09 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Synthesis and evaluation of sulfonyl piperazine LpxH inhibitors.
Bioorg.Chem., 102, 2020
|
|
4R3R
 
 | | Crystal structures of EGFR in complex with Mig6 | | Descriptor: | Epidermal growth factor receptor, peptide from ERBB receptor feedback inhibitor 1' | | Authors: | Park, E, Kim, N, Yi, Z, Cho, A, Kim, K, Ficarro, S.B, Park, A, Park, W.Y, Murray, B, Meyerson, M, Beroukim, R, Marto, J.A, Cho, J, Eck, M.J. | | Deposit date: | 2014-08-17 | | Release date: | 2015-08-12 | | Last modified: | 2015-09-16 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure and mechanism of activity-based inhibition of the EGF receptor by Mig6.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4R3P
 
 | | Crystal structures of EGFR in complex with Mig6 | | Descriptor: | Epidermal growth factor receptor, peptide from ERBB receptor feedback inhibitor 1 | | Authors: | Park, E, Kim, N, Yi, Z, Cho, A, Kim, K, Ficarro, S.B, Park, A, Park, W.Y, Murray, B, Meyerson, M, Beroukim, R, Marto, J.A, Cho, J, Eck, M.J. | | Deposit date: | 2014-08-17 | | Release date: | 2015-08-12 | | Last modified: | 2015-09-16 | | Method: | X-RAY DIFFRACTION (2.905 Å) | | Cite: | Structure and mechanism of activity-based inhibition of the EGF receptor by Mig6.
Nat.Struct.Mol.Biol., 22, 2015
|
|
