3BUE
 
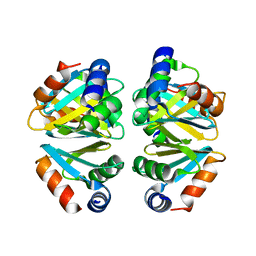 | | Crystal structure of the C-terminal domain hexamer of ArgR from Mycobacterium tuberculosis | | Descriptor: | Arginine repressor ArgR | | Authors: | Cherney, L.T, Cherney, M.M, Garen, C.R, Lu, G.J, James, M.N.G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2008-01-02 | | Release date: | 2008-01-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of the C-terminal domain of the arginine repressor protein from Mycobacterium tuberculosis.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
1NFP
 
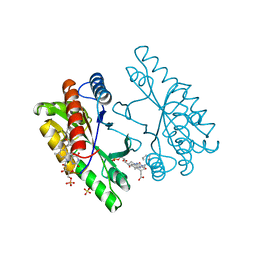 | |
4H88
 
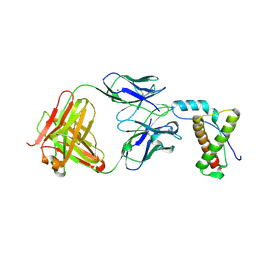 | | Structure of POM1 FAB fragment complexed with mouse PrPc Fragment 120-230 | | Descriptor: | Major prion protein, POM1 FAB CHAIN H, POM1 FAB CHAIN L, ... | | Authors: | Baral, P.K, Wieland, B, Swayampakula, M, James, M.N. | | Deposit date: | 2012-09-21 | | Release date: | 2013-07-31 | | Last modified: | 2013-09-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The toxicity of antiprion antibodies is mediated by the flexible tail of the prion protein.
Nature, 501, 2013
|
|
1S2B
 
 | | Structure of SCP-B the first member of the Eqolisin family of Peptidases to have its structure determined | | Descriptor: | Scytalidopepsin B | | Authors: | Fujinaga, M, Cherney, M.M, Oyama, H, Oda, K, James, M.N. | | Deposit date: | 2004-01-08 | | Release date: | 2004-04-27 | | Last modified: | 2011-11-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The molecular structure and catalytic mechanism of a novel carboxyl peptidase from Scytalidium lignicolum
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1S2K
 
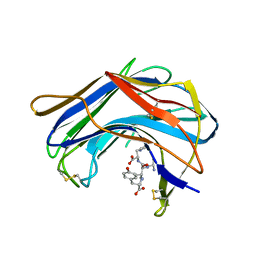 | | Structure of SCP-B a member of the Eqolisin family of Peptidases in a complex with a Tripeptide Ala-Ile-His | | Descriptor: | Ala-Ile-His tripeptide, Scytalidopepsin B, TYROSINE | | Authors: | Fujinaga, M, Cherney, M.M, Oyama, H, Oda, K, James, M.N. | | Deposit date: | 2004-01-08 | | Release date: | 2004-04-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The molecular structure and catalytic mechanism of a novel carboxyl peptidase from Scytalidium lignicolum
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
3IT4
 
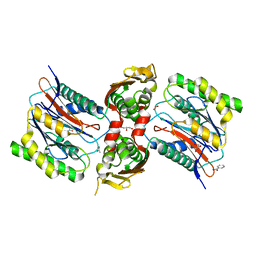 | | The Crystal Structure of Ornithine Acetyltransferase from Mycobacterium tuberculosis (Rv1653) at 1.7 A | | Descriptor: | ACETATE ION, Arginine biosynthesis bifunctional protein argJ alpha chain, Arginine biosynthesis bifunctional protein argJ beta chain, ... | | Authors: | Sankaranarayanan, R, Cherney, M.M, Garen, C, Garen, G, Yuan, M, James, M.N, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2009-08-27 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The molecular structure of ornithine acetyltransferase from Mycobacterium tuberculosis bound to ornithine, a competitive inhibitor.
J.Mol.Biol., 397, 2010
|
|
3IT6
 
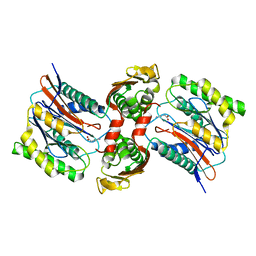 | | The Crystal Structure of Ornithine Acetyltransferase complexed with Ornithine from Mycobacterium tuberculosis (Rv1653) at 2.4 A | | Descriptor: | Arginine biosynthesis bifunctional protein argJ alpha chain, Arginine biosynthesis bifunctional protein argJ beta chain, L-ornithine | | Authors: | Sankaranarayanan, R, Cherney, M.M, Garen, C, Garen, G, Yuan, M, James, M.N, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2009-08-27 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The molecular structure of ornithine acetyltransferase from Mycobacterium tuberculosis bound to ornithine, a competitive inhibitor.
J.Mol.Biol., 397, 2010
|
|
3KPK
 
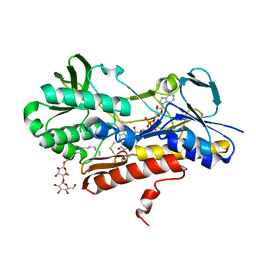 | | Crystal structure of sulfide:quinone oxidoreductase from Acidithiobacillus ferrooxidans, C160A mutant | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FLAVIN-ADENINE DINUCLEOTIDE, HYDROSULFURIC ACID, ... | | Authors: | Cherney, M.M, Zhang, Y, Solomonson, M, Weiner, J.H, James, M.N.G. | | Deposit date: | 2009-11-16 | | Release date: | 2010-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of sulfide:quinone oxidoreductase from Acidithiobacillus ferrooxidans: insights into sulfidotrophic respiration and detoxification.
J.Mol.Biol., 398, 2010
|
|
4J8R
 
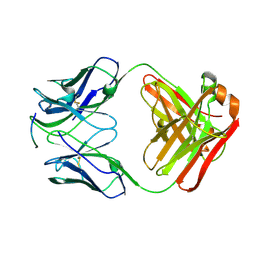 | | Structure of an octapeptide repeat of the prion protein bound to the POM2 Fab antibody fragment | | Descriptor: | Heavy chain of POM2 Fab, Light chain of POM2 Fab, Major prion protein | | Authors: | Swayampakula, M, Baral, P.K, Kav, N.N.V, Aguzzi, A, James, M.N.G. | | Deposit date: | 2013-02-14 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | The crystal structure of an octapeptide repeat of the Prion protein in complex with a Fab fragment of the POM2 antibody.
Protein Sci., 22, 2013
|
|
3LPN
 
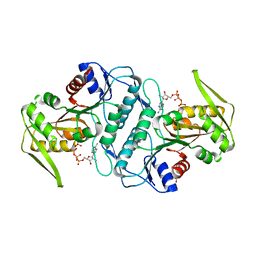 | | Crystal structure of the phosphoribosylpyrophosphate (PRPP) synthetase from Thermoplasma volcanium in complex with an ATP analog (AMPCPP). | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, Ribose-phosphate pyrophosphokinase, SULFATE ION | | Authors: | Cherney, M.M, Cherney, L.T, Garen, C.R, James, M.N.G. | | Deposit date: | 2010-02-05 | | Release date: | 2011-02-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structures of Thermoplasma volcanium phosphoribosyl pyrophosphate synthetase bound to ribose-5-phosphate and ATP analogs.
J.Mol.Biol., 413, 2011
|
|
3LAP
 
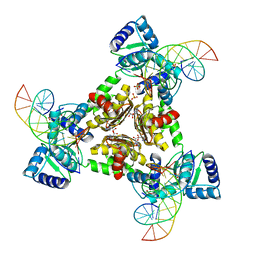 | | The Structure of the Intermediate Complex of the Arginine Repressor from Mycobacterium tuberculosis Bound to its DNA Operator and L-canavanine. | | Descriptor: | 5'-D(*TP*TP*GP*CP*AP*TP*AP*AP*CP*GP*AP*TP*GP*CP*AP*A)-3', 5'-D(*TP*TP*GP*CP*AP*TP*CP*GP*TP*TP*AP*TP*GP*CP*AP*A)-3', Arginine repressor, ... | | Authors: | Cherney, L.T, Cherney, M.M, Garen, C.R, James, M.N.G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-01-06 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | crystal structure of the intermediate complex of the arginine repressor from Mycobacterium tuberculosis bound with its DNA operator reveals detailed mechanism of arginine repression.
J.Mol.Biol., 399, 2010
|
|
3LAJ
 
 | | The Structure of the Intermediate Complex of the Arginine Repressor from Mycobacterium tuberculosis Bound to its DNA Operator and L-arginine. | | Descriptor: | 5'-D(*TP*TP*GP*CP*AP*TP*AP*AP*CP*GP*AP*TP*GP*CP*AP*A)-3', 5'-D(*TP*TP*GP*CP*AP*TP*CP*GP*TP*TP*AP*TP*GP*CP*AP*A)-3', ARGININE, ... | | Authors: | Cherney, L.T, Cherney, M.M, Garen, C.R, James, M.N.G, Mycobacterium Tuberculosis Structural Proteomics Project (XMTB) | | Deposit date: | 2010-01-06 | | Release date: | 2010-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.306 Å) | | Cite: | crystal structure of the intermediate complex of the arginine repressor from Mycobacterium tuberculosis bound with its DNA operator reveals detailed mechanism of arginine repression.
J.Mol.Biol., 399, 2010
|
|
3LRT
 
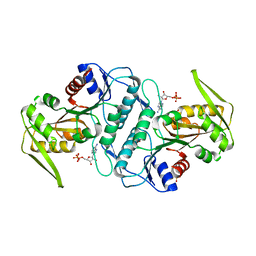 | | Crystal structure of the phosphoribosyl pyrophosphate (PRPP) synthetase from Thermoplasma volcanium in complex with ADP. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Ribose-phosphate pyrophosphokinase, SULFATE ION | | Authors: | Cherney, M.M, Cherney, L.T, Garen, C.R, James, M.N.G. | | Deposit date: | 2010-02-11 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.534 Å) | | Cite: | The structures of Thermoplasma volcanium phosphoribosyl pyrophosphate synthetase bound to ribose-5-phosphate and ATP analogs.
J.Mol.Biol., 413, 2011
|
|
4KGJ
 
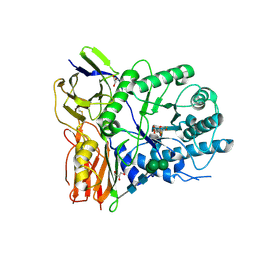 | | Crystal structure of human alpha-L-iduronidase complex with 5-fluoro-alpha-L-idopyranosyluronic acid fluoride | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-fluoro-alpha-L-idopyranosyluronic acid fluoride, ... | | Authors: | Bie, H, Yin, J, He, X, Kermode, A.R, Goddard-Borger, E.D, Withers, S.G, James, M.N.G. | | Deposit date: | 2013-04-29 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Insights into mucopolysaccharidosis I from the structure and action of alpha-L-iduronidase.
Nat.Chem.Biol., 9, 2013
|
|
4KH2
 
 | | Crystal structure of human alpha-L-iduronidase complex with 2-deoxy-2-fluoro-alpha-L-idopyranosyluronic acid fluoride | | Descriptor: | 2,6-anhydro-5-deoxy-5-fluoro-L-idonic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bie, H, Yin, J, He, X, Kermode, A.R, Goddard-Borger, E.D, Withers, S.G, James, M.N.G. | | Deposit date: | 2013-04-29 | | Release date: | 2013-09-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Insights into mucopolysaccharidosis I from the structure and action of alpha-L-iduronidase.
Nat.Chem.Biol., 9, 2013
|
|
1JAK
 
 | | Streptomyces plicatus beta-N-acetylhexosaminidase in Complex with (2R,3R,4S,5R)-2-acetamido-3,4-dihydroxy-5-hydroxymethyl-piperidinium chloride (IFG) | | Descriptor: | (2R,3R,4S,5R)-2-ACETAMIDO-3,4-DIHYDROXY-5-HYDROXYMETHYL-PIPERIDINE, Beta-N-acetylhexosaminidase, CHLORIDE ION, ... | | Authors: | Mark, B.L, Vocadlo, D.J, Zhao, D, Knapp, S, Withers, S.G, James, M.N. | | Deposit date: | 2001-05-30 | | Release date: | 2001-11-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Biochemical and structural assessment of the 1-N-azasugar GalNAc-isofagomine as a potent family 20 beta-N-acetylhexosaminidase inhibitor.
J.Biol.Chem., 276, 2001
|
|
1BXO
 
 | | ACID PROTEINASE (PENICILLOPEPSIN) (E.C.3.4.23.20) COMPLEX WITH PHOSPHONATE INHIBITOR: METHYL CYCLO[(2S)-2-[[(1R)-1-(N-(L-N-(3-METHYLBUTANOYL)VALYL-L-ASPARTYL)AMINO)-3-METHYLBUT YL] HYDROXYPHOSPHINYLOXY]-3-(3-AMINOMETHYL) PHENYLPROPANOATE | | Descriptor: | GLYCEROL, METHYL CYCLO[(2S)-2-[[(1R)-1-(N-(L-N-(3-METHYLBUTANOYL)VALYL-L-ASPARTYL)AMINO)-3-METHYLBUTYL]HYDROXYPHOSPHINYLOXY]-3-(3-AMINOMETHYL)PHENYLPROPANOATE, PROTEIN (PENICILLOPEPSIN), ... | | Authors: | Khan, A.R, Parrish, J.C, Fraser, M.E, Smith, W.W, Bartlett, P.A, James, M.N.G. | | Deposit date: | 1998-10-07 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Lowering the entropic barrier for binding conformationally flexible inhibitors to enzymes.
Biochemistry, 37, 1998
|
|
1BXQ
 
 | | ACID PROTEINASE (PENICILLOPEPSIN) COMPLEX WITH PHOSPHONATE INHIBITOR. | | Descriptor: | 2-[(1R)-1-(N-(3-METHYLBUTANOYL)-L-VALYL-L-ASPARAGINYL)-AMINO)-3-METHYLBUTYL]HYDROXYPHOSPHINYLOXY]-3-PHENYLPROPANOIC ACID METHYLESTER, ACETATE ION, GLYCEROL, ... | | Authors: | Parrish, J.C, Khan, A.R, Fraser, M.E, Smith, W.W, Bartlett, P.A, James, M.N.G. | | Deposit date: | 1998-10-07 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Lowering the entropic barrier for binding conformationally flexible inhibitors to enzymes.
Biochemistry, 37, 1998
|
|
2H6M
 
 | | An episulfide cation (thiiranium ring) trapped in the active site of HAV 3C proteinase inactivated by peptide-based ketone inhibitors | | Descriptor: | N-ACETYL-LEUCYL-ALANYL-ALANYL-(N,N-DIMETHYL)-GLUTAMINE-(1,4-DIOXO-3,4-DIHYDRO-1H-PHTHALAZIN-2-YL)METHYLKETONE INHIBITOR, N-[(BENZYLOXY)CARBONYL]-L-ALANINE, Picornain 3C | | Authors: | Yin, J, Cherney, M.M, Bergmann, E.M, James, M.N. | | Deposit date: | 2006-05-31 | | Release date: | 2006-08-08 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An episulfide cation (thiiranium ring) trapped in the active site of HAV 3C proteinase inactivated by peptide-based ketone inhibitors.
J.Mol.Biol., 361, 2006
|
|
9LYZ
 
 | |
4YX2
 
 | | Crystal structure of Bovine prion protein complexed with POM1 FAB | | Descriptor: | Major prion protein, POM1 FAB HEAVY CHAIN, POM1 FAB LIGHT CHAIN | | Authors: | Baral, P.K, Swayampakula, M, James, M.N.G. | | Deposit date: | 2015-03-22 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | X-ray structural and molecular dynamical studies of the globular domains of cow, deer, elk and Syrian hamster prion proteins.
J.Struct.Biol., 192, 2015
|
|
4YXL
 
 | | Crystal structure of Syrian hamster prion protein complexed with POM1 FAB | | Descriptor: | Major prion protein, POM1 FAB HEAVY CHAIN, POM1 FAB LIGHT CHAIN, ... | | Authors: | Baral, P.K, Swayampakula, M, James, M.N.G. | | Deposit date: | 2015-03-23 | | Release date: | 2015-09-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | X-ray structural and molecular dynamical studies of the globular domains of cow, deer, elk and Syrian hamster prion proteins.
J.Struct.Biol., 192, 2015
|
|
4YXK
 
 | | Crystal structure of Elk prion protein complexed with POM1 FAB | | Descriptor: | Major prion protein, POM1 FAB HEAVY CHAIN, POM1 FAB LIGHT CHAIN, ... | | Authors: | Baral, P.K, Swayampakula, M, James, M.N.G. | | Deposit date: | 2015-03-23 | | Release date: | 2015-09-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.805 Å) | | Cite: | X-ray structural and molecular dynamical studies of the globular domains of cow, deer, elk and Syrian hamster prion proteins.
J.Struct.Biol., 192, 2015
|
|
4YXH
 
 | | Crystal structure of Deer prion protein complexed with POM1 FAB | | Descriptor: | Major prion protein, POM1 FAB HEAVY CHAIN, POM1 FAB LIGHT CHAIN, ... | | Authors: | Baral, P.K, Swayampakula, M, James, M.N.G. | | Deposit date: | 2015-03-23 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | X-ray structural and molecular dynamical studies of the globular domains of cow, deer, elk and Syrian hamster prion proteins.
J.Struct.Biol., 192, 2015
|
|
1CT2
 
 | | CRYSTAL STRUCTURE OF THE OMTKY3 P1 VARIANT OMTKY3-THR18I IN COMPLEX WITH SGPB | | Descriptor: | OVOMUCOID INHIBITOR, PROTEINASE B | | Authors: | Bateman, K.S, Anderson, S, Lu, W, Qasim, M.A, Laskowski Jr, M, James, M.N. | | Deposit date: | 1999-08-18 | | Release date: | 2000-01-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Deleterious effects of beta-branched residues in the S1 specificity pocket of Streptomyces griseus proteinase B (SGPB): crystal structures of the turkey ovomucoid third domain variants Ile18I, Val18I, Thr18I, and Ser18I in complex with SGPB.
Protein Sci., 9, 2000
|
|
