5R5N
 
 | | PanDDA analysis group deposition -- Crystal Structure of human NUDT22 in complex with N13595a | | Descriptor: | 4-chloro-N-cyclopentyl-1-methyl-1H-pyrazole-3-carboxamide, DIMETHYL SULFOXIDE, Uridine diphosphate glucose pyrophosphatase NUDT22 | | Authors: | Diaz-Saez, L, Talon, R, Krojer, T, Burgess-Brown, N.A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Huber, K.V.M. | | Deposit date: | 2020-02-28 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5R64
 
 | | PanDDA analysis group deposition -- Crystal Structure of HUMAN CLEAVAGE FACTOR IM in complex with FMOPL000464a | | Descriptor: | ACETATE ION, Cleavage and polyadenylation specificity factor subunit 5, N-[(2S)-2-hydroxypropyl]-N'-phenylurea, ... | | Authors: | Talon, R, Krojer, T, Diaz-Saez, L, Bradley, A.R, Aimon, A, Fairhead, M, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Huber, K.V.M, von Delft, F. | | Deposit date: | 2020-02-28 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5R5B
 
 | | PanDDA analysis group deposition -- Crystal Structure of human NUDT22 in complex with N13725a | | Descriptor: | 4-[(phenylmethyl)amino]benzoic acid, DIMETHYL SULFOXIDE, Uridine diphosphate glucose pyrophosphatase NUDT22 | | Authors: | Diaz-Saez, L, Talon, R, Krojer, T, Burgess-Brown, N.A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Huber, K.V.M. | | Deposit date: | 2020-02-28 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5R5M
 
 | | PanDDA analysis group deposition -- Crystal Structure of human NUDT22 in complex with N14123a | | Descriptor: | (1-methylpiperidin-4-yl) 3-fluoranylbenzoate, DIMETHYL SULFOXIDE, Uridine diphosphate glucose pyrophosphatase NUDT22 | | Authors: | Diaz-Saez, L, Talon, R, Krojer, T, Burgess-Brown, N.A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Huber, K.V.M. | | Deposit date: | 2020-02-28 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5R50
 
 | | PanDDA analysis group deposition -- Crystal Structure of human NUDT22 in complex with N13371a | | Descriptor: | DIMETHYL SULFOXIDE, N-[4-(aminomethyl)phenyl]methanesulfonamide, Uridine diphosphate glucose pyrophosphatase NUDT22 | | Authors: | Diaz-Saez, L, Talon, R, Krojer, T, Burgess-Brown, N.A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Huber, K.V.M. | | Deposit date: | 2020-02-28 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5R5I
 
 | | PanDDA analysis group deposition -- Crystal Structure of human NUDT22 in complex with N06325b | | Descriptor: | 1-ethanoylpiperidine-4-carbonitrile, DIMETHYL SULFOXIDE, Uridine diphosphate glucose pyrophosphatase NUDT22 | | Authors: | Diaz-Saez, L, Talon, R, Krojer, T, Burgess-Brown, N.A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Huber, K.V.M. | | Deposit date: | 2020-02-28 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
4C9W
 
 | | Crystal structure of NUDT1 (MTH1) with R-crizotinib | | Descriptor: | 3-[(1R)-1-(2,6-dichloro-3-fluorophenyl)ethoxy]-5-(1-piperidin-4-yl-1H-pyrazol-4-yl)pyridin-2-amine, 7,8-DIHYDRO-8-OXOGUANINE TRIPHOSPHATASE, CHLORIDE ION, ... | | Authors: | Elkins, J.M, Salah, E, Huber, K, Superti-Furga, G, Abdul Azeez, K.R, Raynor, J, Krojer, T, von Delft, F, Bountra, C, Edwards, A, Knapp, S. | | Deposit date: | 2013-10-03 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Stereospecific Targeting of Mth1 by (S)-Crizotinib as an Anticancer Strategy.
Nature, 508, 2014
|
|
4C9X
 
 | | Crystal structure of NUDT1 (MTH1) with S-crizotinib | | Descriptor: | 3-[(1S)-1-(2,6-DICHLORO-3-FLUOROPHENYL)ETHOXY]-5-(1-PIPERIDIN-4-YLPYRAZOL-4-YL)PYRIDIN-2-AMINE, 7,8-DIHYDRO-8-OXOGUANINE TRIPHOSPHATASE, CHLORIDE ION, ... | | Authors: | Elkins, J.M, Salah, E, Huber, K, Superti-Furga, G, Abdul Azeez, K.R, Krojer, T, von Delft, F, Bountra, C, Edwards, A, Knapp, S. | | Deposit date: | 2013-10-03 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Stereospecific Targeting of Mth1 by (S)-Crizotinib as an Anticancer Strategy.
Nature, 508, 2014
|
|
6Q3X
 
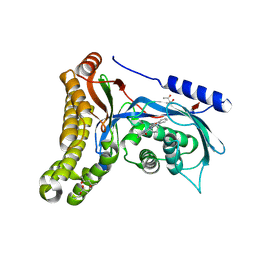 | | Structure of human galactokinase in complex with galactose and 2'-(benzo[d]oxazol-2-ylamino)-7',8'-dihydro-1'H-spiro[cyclohexane-1,4'-quinazolin]-5'(6'H)-one | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(1,3-benzoxazol-2-ylamino)spiro[1,6,7,8-tetrahydroquinazoline-4,1'-cyclohexane]-5-one, Galactokinase, ... | | Authors: | Bezerra, G.A, Mackinnon, S, Zhang, M, Foster, W, Bailey, H, Arrowsmith, C, Edwards, A, Bountra, C, Lai, K, Yue, W.W, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-04 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Fragment Screening Reveals Starting Points for Rational Design of Galactokinase 1 Inhibitors to Treat Classic Galactosemia.
Acs Chem.Biol., 16, 2021
|
|
8OTV
 
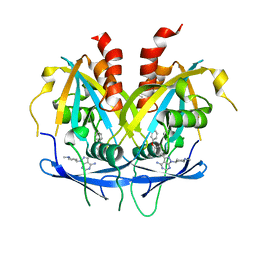 | | Crystal structure of NUDT14 complexed with novel compound | | Descriptor: | 1-(1-methylpiperidin-4-yl)-3-(4-phenoxyphenyl)pyrazolo[3,4-d]pyrimidin-4-amine, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Balikci, E, Feyerherm, C, Bradshaw, W, Seupel, R, Brennan, P.E, Bountra, C, von Delft, F, Huber, K, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-04-21 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Unexpected Noncovalent Off-Target Activity of Clinical BTK Inhibitors Leads to Discovery of a Dual NUDT5/14 Antagonist.
J.Med.Chem., 67, 2024
|
|
6Y6Z
 
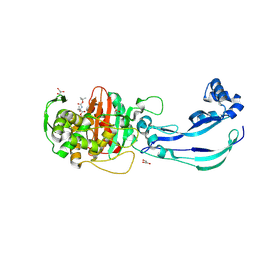 | | Structure of Pseudomonas aeruginosa Penicillin-Binding Protein 3 (PBP3) in complex with Compound 1 | | Descriptor: | GLYCEROL, Peptidoglycan D,D-transpeptidase FtsI, ~{tert}-butyl ~{N}-[(2~{S})-2-methyl-4-oxidanyl-1-oxidanylidene-pent-4-en-2-yl]carbamate | | Authors: | Newman, H, Bellini, D, Dowson, C.G. | | Deposit date: | 2020-02-27 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Demonstration of the utility of DOS-derived fragment libraries for rapid hit derivatisation in a multidirectional fashion.
Chem Sci, 11, 2020
|
|
8AG2
 
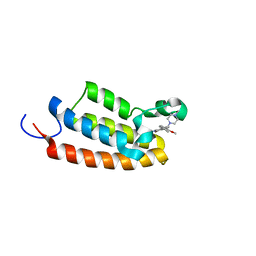 | | Crystal structure of the BPTF bromodomain in complex with BI-7190 | | Descriptor: | 5-[3-methoxy-4-[1-(4-methylpiperazin-1-yl)cyclopropyl]phenyl]-1,3,4-trimethyl-pyridin-2-one, Nucleosome-remodeling factor subunit BPTF | | Authors: | Bader, G, Boettcher, J, Wolkerstorfer, B. | | Deposit date: | 2022-07-19 | | Release date: | 2022-08-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.025 Å) | | Cite: | Discovery of a Chemical Probe to Study Implications of BPTF Bromodomain Inhibition in Cellular and in vivo Experiments.
Chemmedchem, 18, 2023
|
|
8AHC
 
 | | Crystal structure of the BRD9 bromodomain with BI-7189 | | Descriptor: | Bromodomain-containing protein 9, [2,6-dimethoxy-4-(1,2,5-trimethyl-6-oxidanylidene-pyridin-3-yl)phenyl]methyl-dimethyl-azanium | | Authors: | Bader, G, Boettcher, J, Weiss-Puxbaum, A. | | Deposit date: | 2022-07-21 | | Release date: | 2023-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.504 Å) | | Cite: | Discovery of a Chemical Probe to Study Implications of BPTF Bromodomain Inhibition in Cellular and in vivo Experiments.
Chemmedchem, 18, 2023
|
|
6HT1
 
 | | Crystal structure of MLLT1 (ENL) YEATS domain in complexed with SGC-iMLLT (compound 92) | | Descriptor: | 1,2-ETHANEDIOL, 1-methyl-~{N}-[2-[[(2~{S})-2-methylpyrrolidin-1-yl]methyl]-3~{H}-benzimidazol-5-yl]indazole-5-carboxamide, Protein ENL, ... | | Authors: | Heidenreich, D, Chaikuad, A, Moustakim, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fedorov, O, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-10-02 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of an MLLT1/3 YEATS Domain Chemical Probe.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6I8L
 
 | | Crystal structure of Spindlin1 in complex with the inhibitor TD001851a | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 5'-(cyclopropylmethoxy)-6'-[3-(1,3-dihydroisoindol-2-yl)propoxy]spiro[cyclopentane-1,3'-indole]-2'-amine, ... | | Authors: | Johansson, C, Fagan, V, Brennan, P.E, Sorrell, F.J, Krojer, T, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U.C.T. | | Deposit date: | 2018-11-20 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | A Chemical Probe for Tudor Domain Protein Spindlin1 to Investigate Chromatin Function.
J.Med.Chem., 62, 2019
|
|
6I8Y
 
 | | Crystal structure of Spindlin1 in complex with the Methyltransferase inhibitor A366 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 5'-methoxy-6'-[3-(pyrrolidin-1-yl)propoxy]spiro[cyclobutane-1,3'-indol]-2'-amine, ... | | Authors: | Srikannathasan, V, Johansson, C, Gileadi, C, Shrestha, L, Sorrell, F.J, Krojer, T, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U.C.T. | | Deposit date: | 2018-11-21 | | Release date: | 2018-12-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | A Chemical Probe for Tudor Domain Protein Spindlin1 to Investigate Chromatin Function.
J.Med.Chem., 62, 2019
|
|
6I8B
 
 | | Crystal structure of Spindlin1 in complex with the inhibitor VinSpinIn | | Descriptor: | 2-[4-[2-[[2-[3-[2-azanyl-5-(cyclopropylmethoxy)-3,3-dimethyl-indol-6-yl]oxypropyl]-1,3-dihydroisoindol-5-yl]oxy]ethyl]-1,2,3-triazol-1-yl]-1-[4-(2-pyrrolidin-1-ylethyl)piperidin-1-yl]ethanone, DIMETHYL SULFOXIDE, GLYCINE, ... | | Authors: | Johansson, C, Fagan, V, Brennan, P.E, Sorrell, F.J, Krojer, T, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U.C.T. | | Deposit date: | 2018-11-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | A Chemical Probe for Tudor Domain Protein Spindlin1 to Investigate Chromatin Function.
J.Med.Chem., 62, 2019
|
|
6HT0
 
 | | Crystal structure of MLLT1 (ENL) YEATS domain in complexed with compound 94 | | Descriptor: | 1,2-ETHANEDIOL, 1-cyclopropyl-~{N}-[2-[[(2~{S})-2-methylpyrrolidin-1-yl]methyl]-3~{H}-benzimidazol-5-yl]indazole-5-carboxamide, Protein ENL, ... | | Authors: | Heidenreich, D, Chaikuad, A, Moustakim, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fedorov, O, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-10-02 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of an MLLT1/3 YEATS Domain Chemical Probe.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6F22
 
 | | Complex between MTH1 and compound 29 (a 4-amino-2,7-diazaindole derivative) | | Descriptor: | (3~{S})-3-phenyl-4-(2~{H}-pyrazolo[3,4-b]pyridin-4-yl)morpholine, 7,8-dihydro-8-oxoguanine triphosphatase, SULFATE ION | | Authors: | Viklund, J, Talagas, A, Tresaugues, L, Andersson, M, Ericsson, U, Forsblom, R, Ginman, T, Hallberg, K, Lindstrom, J, Persson, L, Silvander, C, Rahm, F. | | Deposit date: | 2017-11-23 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Creation of a Novel Class of Potent and Selective MutT Homologue 1 (MTH1) Inhibitors Using Fragment-Based Screening and Structure-Based Drug Design.
J. Med. Chem., 61, 2018
|
|
6F1X
 
 | | Complex between MTH1 and compound 7 (a 7-azaindole-2-amide derivative) | | Descriptor: | 4-(3-chlorophenyl)-~{N}-ethyl-1~{H}-pyrrolo[2,3-b]pyridine-2-carboxamide, 7,8-dihydro-8-oxoguanine triphosphatase, SULFATE ION | | Authors: | Viklund, J, Talagas, A, Tresaugues, L, Andersson, M, Ericsson, U, Forsblom, R, Ginman, T, Hallberg, K, Lindstrom, J, Persson, L, Silvander, C, Rahm, F. | | Deposit date: | 2017-11-23 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Creation of a Novel Class of Potent and Selective MutT Homologue 1 (MTH1) Inhibitors Using Fragment-Based Screening and Structure-Based Drug Design.
J. Med. Chem., 61, 2018
|
|
6F23
 
 | | Complex between MTH1 and compound 16 (a 4-amino-7-azaindole derivative) | | Descriptor: | 4-[(2~{R})-2-phenylpyrrolidin-1-yl]-1~{H}-pyrrolo[2,3-b]pyridine, 7,8-dihydro-8-oxoguanine triphosphatase, GLYCEROL, ... | | Authors: | Viklund, J, Tresaugues, L, Talagas, A, Andersson, M, Ericsson, U, Forsblom, R, Ginman, T, Hallberg, K, Lindstrom, J, Persson, L, Silvander, C, Rahm, F. | | Deposit date: | 2017-11-23 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Creation of a Novel Class of Potent and Selective MutT Homologue 1 (MTH1) Inhibitors Using Fragment-Based Screening and Structure-Based Drug Design.
J. Med. Chem., 61, 2018
|
|
6F20
 
 | | Complex between MTH1 and compound 1 (a 7-azaindole-4-ester derivative) | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, ACETATE ION, Ethyl 1H-pyrrolo[2,3-b]pyridine-4-carboxylate, ... | | Authors: | Viklund, J, Talagas, A, Tresaugues, L, Andersson, M, Ericsson, U, Forsblom, R, Ginman, T, Hallberg, K, Lindstrom, J, Persson, L, Silvander, C, Rahm, F. | | Deposit date: | 2017-11-23 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Creation of a Novel Class of Potent and Selective MutT Homologue 1 (MTH1) Inhibitors Using Fragment-Based Screening and Structure-Based Drug Design.
J. Med. Chem., 61, 2018
|
|
6G24
 
 | | X-ray structure of NSD3-PWWP1 in complex with compound 3 | | Descriptor: | 2-[(~{E})-2-thiophen-2-ylethenyl]benzoic acid, Histone-lysine N-methyltransferase NSD3 | | Authors: | Boettcher, J, Muellauer, B.J, Weiss-Puxbaum, A, Zoephel, A. | | Deposit date: | 2018-03-22 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Fragment-based discovery of a chemical probe for the PWWP1 domain of NSD3.
Nat.Chem.Biol., 15, 2019
|
|
6G2C
 
 | | X-ray structure of NSD3-PWWP1 in complex with compound 9 | | Descriptor: | 3,5-dimethyl-4-(1-methyl-5-pyridin-4-yl-imidazol-4-yl)-1,2-oxazole, Histone-lysine N-methyltransferase NSD3 | | Authors: | Boettcher, J, Muellauer, B.J, Weiss-Puxbaum, A, Zoephel, A. | | Deposit date: | 2018-03-22 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Fragment-based discovery of a chemical probe for the PWWP1 domain of NSD3.
Nat.Chem.Biol., 15, 2019
|
|
6G2O
 
 | | X-ray structure of NSD3-PWWP1 in complex with compound BI-9321 | | Descriptor: | Histone-lysine N-methyltransferase NSD3, [4-[5-(7-fluoranylquinolin-4-yl)-1-methyl-imidazol-4-yl]-3,5-dimethyl-phenyl]methanamine | | Authors: | Boettcher, J, Muellauer, B.J, Weiss-Puxbaum, A, Zoephel, A. | | Deposit date: | 2018-03-23 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Fragment-based discovery of a chemical probe for the PWWP1 domain of NSD3.
Nat.Chem.Biol., 15, 2019
|
|
