8JHY
 
 | | Cryo-EM structure of compound 9n bound ketone body receptor HCAR2-Gi signaling complex | | Descriptor: | 7-methyl-N-[(2R)-1-phenoxypropan-2-yl]-3-(4-propan-2-ylphenyl)pyrazolo[1,5-a]pyrimidine-6-carboxamide, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhao, C, Tian, X.W, Liu, Y, Cheng, L, Yan, W, Shao, Z.H. | | Deposit date: | 2023-05-25 | | Release date: | 2023-09-06 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Biased allosteric activation of ketone body receptor HCAR2 suppresses inflammation.
Mol.Cell, 83, 2023
|
|
8JIM
 
 | | Cryo-EM structure of MMF bound ketone body receptor HCAR2-Gi signaling complex | | Descriptor: | (2Z)-4-methoxy-4-oxobut-2-enoic acid, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhao, C, Tian, X.W, Liu, Y, Cheng, L, Yan, W, Shao, Z.H. | | Deposit date: | 2023-05-26 | | Release date: | 2023-09-06 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Biased allosteric activation of ketone body receptor HCAR2 suppresses inflammation.
Mol.Cell, 83, 2023
|
|
8JIF
 
 | | Cryo-EM Structure of 3-axis block of AAV9P31-Car4 complex | | Descriptor: | Capsid protein VP1, Carbonic anhydrase 4, ZINC ION | | Authors: | Zhang, R, Liu, Y, Lou, Z. | | Deposit date: | 2023-05-26 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.28 Å) | | Cite: | Structural basis of the recognition of adeno-associated virus by the neurological system-related receptor carbonic anhydrase IV.
Plos Pathog., 20, 2024
|
|
7X8O
 
 | | NMR Solution Structure of the 2:1 Coptisine-KRAS-G4 Complex | | Descriptor: | 6,7-dihydro[1,3]dioxolo[4,5-g][1,3]dioxolo[7,8]isoquino[3,2-a]isoquinolin-5-ium, DNA (24-MER) | | Authors: | Wang, K.B, Liu, Y, Li, J, Xiao, C, Gu, W, Li, Y, Xia, Y.Z, Yan, T, Yang, M.H, Kong, L.Y. | | Deposit date: | 2022-03-14 | | Release date: | 2022-09-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural insight into the bulge-containing KRAS oncogene promoter G-quadruplex bound to berberine and coptisine.
Nat Commun, 13, 2022
|
|
7X8N
 
 | | NMR Solution Structure of the Wild-type Bulge-containing KRAS-G4 | | Descriptor: | DNA (24-mer) | | Authors: | Wang, K.B, Liu, Y, Li, J, Xiao, C, Gu, W, Li, Y, Xia, Y.Z, Yan, T, Yang, M.H, Kong, L.Y. | | Deposit date: | 2022-03-14 | | Release date: | 2022-09-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural insight into the bulge-containing KRAS oncogene promoter G-quadruplex bound to berberine and coptisine.
Nat Commun, 13, 2022
|
|
7WGW
 
 | | NMR Solution Structure of a cGMP Fill-in Vacancy G-quadruplex Formed in the Oxidized BLM Gene Promoter | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, DNA (20-MER) | | Authors: | Wang, K.B, Liu, Y, Li, Y, Li, J, Dickerhoff, J, Yang, M.H, Yang, D, Kong, L.Y. | | Deposit date: | 2021-12-29 | | Release date: | 2022-05-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Oxidative Damage Induces a Vacancy G-Quadruplex That Binds Guanine Metabolites: Solution Structure of a cGMP Fill-in Vacancy G-Quadruplex in the Oxidized BLM Gene Promoter.
J.Am.Chem.Soc., 144, 2022
|
|
7WI7
 
 | | Crystal structure of human MCM8/9 complex | | Descriptor: | DNA helicase MCM8, DNA helicase MCM9, ZINC ION | | Authors: | Li, J, Liu, Y. | | Deposit date: | 2022-01-03 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (6.6 Å) | | Cite: | Crystal structure of human MCM8/9 complex
To Be Published
|
|
7DQO
 
 | | Marsupenaeus japonicus ferritin mutant-D132R | | Descriptor: | FE (III) ION, Ferritin | | Authors: | Tan, X, Liu, Y, Zang, J, Zhang, T, Zhao, G. | | Deposit date: | 2020-12-24 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Hyperthermostability of prawn ferritin nanocage facilitates its application as a robust nanovehicle for nutraceuticals.
Int.J.Biol.Macromol., 191, 2021
|
|
7DQP
 
 | | Thermal treated Marsupenaeus japonicus ferritin | | Descriptor: | Ferritin | | Authors: | Tan, X, Liu, Y, Zang, J, Zhang, T, Zhao, G. | | Deposit date: | 2020-12-24 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Hyperthermostability of prawn ferritin nanocage facilitates its application as a robust nanovehicle for nutraceuticals.
Int.J.Biol.Macromol., 191, 2021
|
|
7E29
 
 | | Crystal Structure of Saccharomyces cerevisiae Ioc4 PWWP domain fused with MBP | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,ISWI one complex protein 4, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Li, J, Smolle, M, Liang, H, Liu, Y. | | Deposit date: | 2021-02-05 | | Release date: | 2022-02-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | H3K36 methylation and DNA-binding both promote Ioc4 recruitment and Isw1b remodeler function.
Nucleic Acids Res., 50, 2022
|
|
7YEH
 
 | | Cryo-EM structure of human OGT-OGA complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Protein O-GlcNAcase, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit, ... | | Authors: | Lu, P, Liu, Y, Yu, H, Gao, H. | | Deposit date: | 2022-07-05 | | Release date: | 2023-07-12 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Cryo-EM structure of human O-GlcNAcylation enzyme pair OGT-OGA complex.
Nat Commun, 14, 2023
|
|
7YEA
 
 | | Human O-GlcNAc transferase Dimer | | Descriptor: | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit | | Authors: | Gao, H, Lu, P, Liu, Y. | | Deposit date: | 2022-07-05 | | Release date: | 2023-07-12 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Cryo-EM structure of human O-GlcNAcylation enzyme pair OGT-OGA complex.
Nat Commun, 14, 2023
|
|
6FKQ
 
 | | THE CRYSTAL STRUCTURE OF A FRAGMENT OF NETRIN-1 IN COMPLEX WITH A FRAGMENT OF DRAXIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Bhowmick, T, Meijers, R. | | Deposit date: | 2018-01-24 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Structural Basis for Draxin-Modulated Axon Guidance and Fasciculation by Netrin-1 through DCC.
Neuron, 97, 2018
|
|
4DVF
 
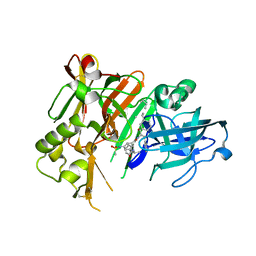 | | Crystal structure of BACE1 with its inhibitor | | Descriptor: | Beta-secretase 1, METHYL (2S)-1-[(2R,5S,8S,12S,13S)-2,13-DIBENZYL-12-HYDROXY-3,5-DIMETHYL-8-(2-METHYLPROPYL)-15-(3-[(METHYLSULFONYL)AMINO]-5-{[(1R)-1-PHENYLETHYL]CARBAMOYL}PHENYL)-4,7,10,15-TETRAOXO-3,6,9,14-TETRAAZAPENTADECAN-1-OYL]PYRROLIDINE-2-CARBOXYLATE | | Authors: | Xu, Y.C, Chen, W.Y, Li, L, Chen, T.T. | | Deposit date: | 2012-02-23 | | Release date: | 2013-01-16 | | Last modified: | 2021-09-15 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Cyanobacterial Peptides as a Prototype for the Design of Potent beta-Secretase Inhibitors and the Development of Selective Chemical Probes for Other Aspartic Proteases
J.Med.Chem., 55, 2012
|
|
5NS3
 
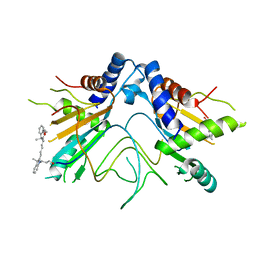 | |
5NS4
 
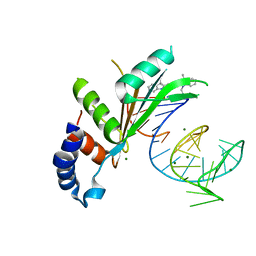 | | Crystal structures of Cy3 cyanine fluorophores stacked onto the end of double-stranded RNA | | Descriptor: | 3-[(2~{Z})-2-[(~{E})-3-[3,3-dimethyl-1-(3-oxidanylpropyl)indol-1-ium-2-yl]prop-2-enylidene]-3,3-dimethyl-indol-1-yl]propan-1-ol, 50S ribosomal protein L5, MAGNESIUM ION, ... | | Authors: | Liu, Y.J, Lilley, D.M.J. | | Deposit date: | 2017-04-25 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of Cyanine Fluorophores Stacked onto the End of Double-Stranded RNA.
Biophys. J., 113, 2017
|
|
7R4H
 
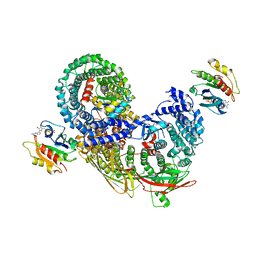 | | phospho-STING binding to adaptor protein complex-1 | | Descriptor: | ADP-ribosylation factor 1, AP-1 complex subunit beta-1, AP-1 complex subunit gamma-1, ... | | Authors: | Xu, P, Ablasser, A. | | Deposit date: | 2022-02-08 | | Release date: | 2022-09-14 | | Last modified: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (2.34 Å) | | Cite: | Clathrin-associated AP-1 controls termination of STING signalling.
Nature, 610, 2022
|
|
6LCG
 
 | | Structure of D-carbamoylase mutant from Nitratireductor indicus | | Descriptor: | DI(HYDROXYETHYL)ETHER, N-carbamoyl-D-amino-acid hydrolase | | Authors: | Liu, Y.F, Ni, Y, Xu, G.C, Dai, W. | | Deposit date: | 2019-11-18 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-Guided Engineering of D-Carbamoylase Reveals a Key Loop at Substrate Entrance Tunnel
Acs Catalysis, 10, 2020
|
|
6LEI
 
 | |
7BWN
 
 | | Crystal Structure of a Designed Protein Heterocatenane | | Descriptor: | Cellular tumor antigen p53, Chimera of Green fluorescent protein and p53dim | | Authors: | Liu, Y.J, Duan, Z.L, Fang, J, Zhang, F, Xiao, J.Y, Zhang, W.B. | | Deposit date: | 2020-04-15 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.396 Å) | | Cite: | Cellular Synthesis and X-ray Crystal Structure of a Designed Protein Heterocatenane.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6LHH
 
 | | Crystal structure of chicken 8mer-BF2*1501 | | Descriptor: | ARG-ARG-ARG-GLU-GLN-THR-ASP-TYR, Beta-2-microglobulin, MHC class I | | Authors: | Liu, Y.J, Xia, C. | | Deposit date: | 2019-12-08 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | The Combination of CD8 alpha alpha and Peptide-MHC-I in a Face-to-Face Mode Promotes Chicken gamma delta T Cells Response.
Front Immunol, 11, 2020
|
|
6LHF
 
 | | Crystal structure of chicken cCD8aa/pBF2*15:01 | | Descriptor: | Beta-2-microglobulin, CD8 alpha chain, MHC class I, ... | | Authors: | Liu, Y.J, Xia, C. | | Deposit date: | 2019-12-08 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Combination of CD8 alpha alpha and Peptide-MHC-I in a Face-to-Face Mode Promotes Chicken gamma delta T Cells Response.
Front Immunol, 11, 2020
|
|
1OQD
 
 | | Crystal structure of sTALL-1 and BCMA | | Descriptor: | Tumor necrosis factor ligand superfamily member 13B, soluble form, Tumor necrosis factor receptor superfamily member 17 | | Authors: | Zhang, G. | | Deposit date: | 2003-03-07 | | Release date: | 2003-05-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Ligand-receptor binding revealed by the TNF family member TALL-1.
Nature, 423, 2003
|
|
1OQE
 
 | | Crystal structure of sTALL-1 with BAFF-R | | Descriptor: | Tumor necrosis factor ligand superfamily member 13B, soluble form, Tumor necrosis factor receptor superfamily member 13C | | Authors: | Zhang, G. | | Deposit date: | 2003-03-07 | | Release date: | 2003-05-13 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ligand-receptor binding revealed by the TNF family member TALL-1.
Nature, 423, 2003
|
|
6LED
 
 | |
