8HC8
 
 | | SARS-CoV-2 Omicron BA.1 spike trimer (6P) in complex with YB13-292 Fab, focused refinement of Fab region | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of YB13-292 Fab, Light chain of YB13-292 Fab, ... | | Authors: | Liu, B, Gao, X, Chen, Q, Li, Z, Su, M, He, J, Xiong, X. | | Deposit date: | 2022-11-01 | | Release date: | 2023-01-25 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | Somatically hypermutated antibodies isolated from SARS-CoV-2 Delta infected patients cross-neutralize heterologous variants.
Nat Commun, 14, 2023
|
|
8HC7
 
 | | SARS-CoV-2 Omicron BA.1 spike trimer (6P) complex with YB9-258 Fab, focused refinement of RBD-dimer region | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain variable region of YB9-258, Light chain variable region of YB9-258, ... | | Authors: | Liu, B, Gao, X, Chen, Q, Li, Z, Su, M, He, J, Xiong, X. | | Deposit date: | 2022-11-01 | | Release date: | 2023-01-25 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (4.54 Å) | | Cite: | Somatically hypermutated antibodies isolated from SARS-CoV-2 Delta infected patients cross-neutralize heterologous variants.
Nat Commun, 14, 2023
|
|
8HCB
 
 | | SARS-CoV-2 Omicron BA.1 spike trimer (6P) in complex with 3 YB13-292 Fabs (2 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of YB13-292 Fab, ... | | Authors: | Liu, B, Gao, X, Chen, Q, Li, Z, Su, M, He, J, Xiong, X. | | Deposit date: | 2022-11-01 | | Release date: | 2023-01-25 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | Somatically hypermutated antibodies isolated from SARS-CoV-2 Delta infected patients cross-neutralize heterologous variants.
Nat Commun, 14, 2023
|
|
8HC3
 
 | | SARS-CoV-2 Omicron BA.1 spike trimer (6P) in complex with 2 YB9-258 Fabs (2 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of YB9-258, ... | | Authors: | Liu, B, Gao, X, Chen, Q, Li, Z, Su, M, He, J, Xiong, X. | | Deposit date: | 2022-11-01 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.35 Å) | | Cite: | Somatically hypermutated antibodies isolated from SARS-CoV-2 Delta infected patients cross-neutralize heterologous variants.
Nat Commun, 14, 2023
|
|
8HC4
 
 | | SARS-CoV-2 wildtype spike trimer (6P) in complex with 3 YB9-258 Fabs and 3 R1-32 Fabs (3 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32 Fab, ... | | Authors: | Liu, B, Gao, X, Chen, Q, Li, Z, Su, M, He, J, Xiong, X. | | Deposit date: | 2022-11-01 | | Release date: | 2023-01-25 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Somatically hypermutated antibodies isolated from SARS-CoV-2 Delta infected patients cross-neutralize heterologous variants.
Nat Commun, 14, 2023
|
|
8HCA
 
 | | SARS-CoV-2 Omicron BA.1 spike trimer (6P) in complex with 3 YB13-292 Fabs (1 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of YB13-292 Fab, ... | | Authors: | Liu, B, Gao, X, Chen, Q, Li, Z, Su, M, He, J, Xiong, X. | | Deposit date: | 2022-11-01 | | Release date: | 2023-01-25 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (4.35 Å) | | Cite: | Somatically hypermutated antibodies isolated from SARS-CoV-2 Delta infected patients cross-neutralize heterologous variants.
Nat Commun, 14, 2023
|
|
8HC6
 
 | | SARS-CoV-2 Omicron BA.1 spike trimer (6P) in complex with YB9-258 Fab, focused refinement of Fab region | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of YB9-258 Fab, Light chain of YB9-258, ... | | Authors: | Liu, B, Gao, X, Chen, Q, Li, Z, Su, M, He, J, Xiong, X. | | Deposit date: | 2022-11-01 | | Release date: | 2023-01-25 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (4.69 Å) | | Cite: | Somatically hypermutated antibodies isolated from SARS-CoV-2 Delta infected patients cross-neutralize heterologous variants.
Nat Commun, 14, 2023
|
|
8KG5
 
 | | Prefusion RSV F Bound to Lonafarnib and D25 Fab | | Descriptor: | 4-{2-[4-(3,10-DIBROMO-8-CHLORO-6,11-DIHYDRO-5H-BENZO[5,6]CYCLOHEPTA[1,2-B]PYRIDIN-11-YL)PIPERIDIN-1-YL]-2-OXOETHYL}PIPERIDINE-1-CARBOXAMIDE, D25 heavy chain, D25 light chain, ... | | Authors: | Yang, Q, Xue, B, Liu, F, Peng, W, Chen, X. | | Deposit date: | 2023-08-17 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Farnesyltransferase inhibitor lonafarnib suppresses respiratory syncytial virus infection by blocking conformational change of fusion glycoprotein.
Signal Transduct Target Ther, 9, 2024
|
|
8HC9
 
 | | SARS-CoV-2 Omicron BA.1 spike trimer (6P) in complex with 3 YB13-292 Fabs (3 RBD down) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of YB13-292 Fab, ... | | Authors: | Liu, B, Gao, X, Chen, Q, Li, Z, Su, M, He, J, Xiong, X. | | Deposit date: | 2022-11-01 | | Release date: | 2023-01-25 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (6.03 Å) | | Cite: | Somatically hypermutated antibodies isolated from SARS-CoV-2 Delta infected patients cross-neutralize heterologous variants.
Nat Commun, 14, 2023
|
|
8HC2
 
 | | SARS-CoV-2 Omicron BA.1 spike trimer (6P) in complex with 1 YB9-258 Fab (1 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of YB9-258 Fab, ... | | Authors: | Liu, B, Gao, X, Chen, Q, Li, Z, Su, M, He, J, Xiong, X. | | Deposit date: | 2022-11-01 | | Release date: | 2023-01-25 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (6.21 Å) | | Cite: | Somatically hypermutated antibodies isolated from SARS-CoV-2 Delta infected patients cross-neutralize heterologous variants.
Nat Commun, 14, 2023
|
|
2G8D
 
 | |
7Y7J
 
 | | SARS-CoV-2 S trimer in complex with 1F Fab | | Descriptor: | 1F VH, 1F VL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, S, Liu, F, Yang, X, Zhong, G. | | Deposit date: | 2022-06-22 | | Release date: | 2023-02-22 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | A core epitope targeting antibody of SARS-CoV-2.
Protein Cell, 14, 2023
|
|
7Y7K
 
 | | SARS-CoV-2 RBD in complex with 1F Fab | | Descriptor: | 1F VH, 1F VL, Spike protein S1 | | Authors: | Zhao, S, Liu, F, Yang, X, Zhong, G. | | Deposit date: | 2022-06-22 | | Release date: | 2023-02-22 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | A core epitope targeting antibody of SARS-CoV-2.
Protein Cell, 14, 2023
|
|
8IT9
 
 | | Co-crystal structure of FTO bound to 22 | | Descriptor: | 2-OXOGLUTARIC ACID, 2-[(2,6-diethyl-4-pyridin-4-yl-phenyl)amino]-6-(1,4-oxazepan-4-ylmethyl)benzoic acid, Alpha-ketoglutarate-dependent dioxygenase FTO | | Authors: | Yang, C.-G, Gan, J.H. | | Deposit date: | 2023-03-22 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Rational Design of RNA Demethylase FTO Inhibitors with Enhanced Antileukemia Drug-Like Properties.
J.Med.Chem., 66, 2023
|
|
6E1A
 
 | | Menin bound to M-89 | | Descriptor: | (1S,2R)-2-[(4S)-2-methyl-4-{1-[(1-{4-[(pyridin-4-yl)sulfonyl]phenyl}azetidin-3-yl)methyl]piperidin-4-yl}-1,2,3,4-tetrahydroisoquinolin-4-yl]cyclopentyl methylcarbamate, Menin, praseodymium triacetate | | Authors: | Stuckey, J.A. | | Deposit date: | 2018-07-09 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure-Based Discovery of M-89 as a Highly Potent Inhibitor of the Menin-Mixed Lineage Leukemia (Menin-MLL) Protein-Protein Interaction.
J.Med.Chem., 62, 2019
|
|
6E8E
 
 | |
6E8D
 
 | |
6LWF
 
 | | Crystal structure of human NEIL1(P2G, E3Q, K242) bound to duplex DNA containing guanidinohydantoin (Gh) | | Descriptor: | DNA (5'-D(*CP*GP*TP*CP*CP*AP*(DGH)P*GP*TP*CP*TP*AP*C)-3'), DNA (5'-D(*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*G)-3'), Endonuclease 8-like 1 | | Authors: | Liu, M.H, Zhang, J, Zhu, C.X, Zhang, X.X, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2020-02-07 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | DNA repair glycosylase hNEIL1 triages damaged bases via competing interaction modes.
Nat Commun, 12, 2021
|
|
6LWC
 
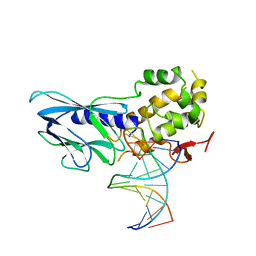 | | Crystal structure of human NEIL1(P2G, E3Q, K242) bound to duplex DNA containing spiroiminodihydantoin (Sp) | | Descriptor: | DNA (5'-D(*CP*GP*TP*CP*CP*AP*(DSP)P*GP*TP*CP*TP*AP*C)-3'), DNA (5'-D(*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*G)-3'), Endonuclease 8-like 1 | | Authors: | Liu, M.H, Zhang, J, Zhu, C.X, Zhang, X.X, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2020-02-07 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | DNA repair glycosylase hNEIL1 triages damaged bases via competing interaction modes.
Nat Commun, 12, 2021
|
|
6LWA
 
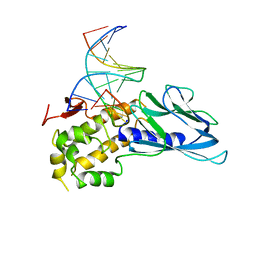 | | Crystal structure of human NEIL1(P2G, E3Q, K242) bound to duplex DNA containing 5-hydroxyuracil (5-OHU) | | Descriptor: | DNA (5'-D(*CP*GP*TP*CP*CP*AP*(OHU)P*GP*TP*CP*TP*AP*C)-3'), DNA (5'-D(*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*G)-3'), Endonuclease 8-like 1 | | Authors: | Liu, M.H, Zhang, J, Zhu, C.X, Zhang, X.X, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2020-02-07 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | DNA repair glycosylase hNEIL1 triages damaged bases via competing interaction modes.
Nat Commun, 12, 2021
|
|
6LWG
 
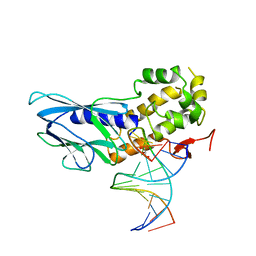 | | Crystal structure of human NEIL1(P2G, E3Q, R242) bound to duplex DNA containing guanidinohydantoin (Gh) | | Descriptor: | DNA (5'-D(*CP*GP*TP*CP*CP*AP*(DGH)P*GP*TP*CP*TP*AP*C)-3'), DNA (5'-D(*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*G)-3'), Endonuclease 8-like 1, ... | | Authors: | Liu, M.H, Zhang, J, Zhu, C.X, Zhang, X.X, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2020-02-07 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | DNA repair glycosylase hNEIL1 triages damaged bases via competing interaction modes.
Nat Commun, 12, 2021
|
|
6LWB
 
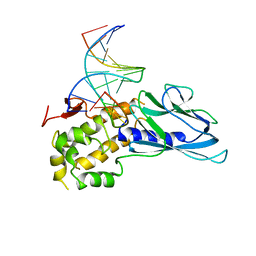 | | Crystal structure of human NEIL1(P2G, E3Q, R242) bound to duplex DNA containing 5-hydroxyuracil (5-OHU) | | Descriptor: | DNA (5'-D(*CP*GP*TP*CP*CP*AP*(OHU)P*GP*TP*CP*TP*AP*C)-3'), DNA (5'-D(*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*G)-3'), Endonuclease 8-like 1 | | Authors: | Liu, M.H, Zhang, J, Zhu, C.X, Zhang, X.X, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2020-02-07 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | DNA repair glycosylase hNEIL1 triages damaged bases via competing interaction modes.
Nat Commun, 12, 2021
|
|
6LWN
 
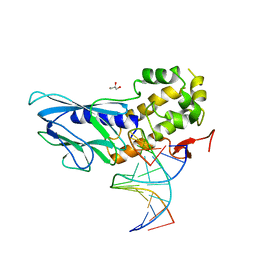 | | Crystal structure of human NEIL1(R242, G249P) bound to duplex DNA containing 2'-fluoro-2'-deoxy-5,6-dihydrouridine | | Descriptor: | DNA (5'-D(*CP*GP*TP*CP*CP*AP*(FDU)P*GP*TP*CP*TP*AP*C)-3'), DNA (5'-D(*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*G)-3'), Endonuclease 8-like 1, ... | | Authors: | Liu, M.H, Zhang, J, Zhu, C.X, Zhang, X.X, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2020-02-07 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | DNA repair glycosylase hNEIL1 triages damaged bases via competing interaction modes.
Nat Commun, 12, 2021
|
|
6LWO
 
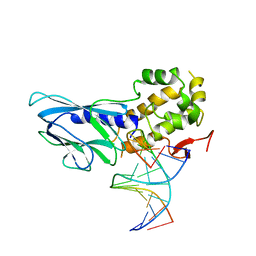 | | Crystal structure of human NEIL1(R242, Y244H) bound to duplex DNA containing 2'-fluoro-2'-deoxy-5,6-dihydrouridine | | Descriptor: | DNA (5'-D(*CP*GP*TP*CP*CP*AP*(FDU)P*GP*TP*CP*TP*AP*C)-3'), DNA (5'-D(*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*G)-3'), Endonuclease 8-like 1, ... | | Authors: | Liu, M.H, Zhang, J, Zhu, C.X, Zhang, X.X, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2020-02-07 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | DNA repair glycosylase hNEIL1 triages damaged bases via competing interaction modes.
Nat Commun, 12, 2021
|
|
6LWM
 
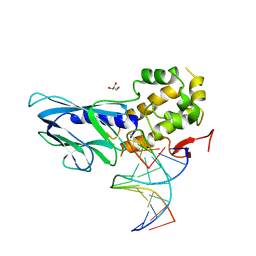 | | Crystal structure of human NEIL1(K242) bound to duplex DNA containing 2'-fluoro-2'-deoxy-5,6-dihydrouridine | | Descriptor: | DNA (5'-D(*CP*GP*TP*CP*CP*AP*(FDU)P*GP*TP*CP*TP*AP*C)-3'), DNA (5'-D(*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*G)-3'), Endonuclease 8-like 1, ... | | Authors: | Liu, M.H, Zhang, J, Zhu, C.X, Zhang, X.X, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2020-02-07 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | DNA repair glycosylase hNEIL1 triages damaged bases via competing interaction modes.
Nat Commun, 12, 2021
|
|
