7KLM
 
 | | Human Arginase1 Complexed with Inhibitor Compound 24a | | Descriptor: | 3-[(2~{S},3~{R},4~{R})-4-[[(2~{S})-2-azanyl-3-methyl-butanoyl]amino]-2-carboxy-pyrrolidin-3-yl]propyl-$l^{3}-oxidanyl-bis(oxidanyl)boron, Arginase-1, MANGANESE (II) ION | | Authors: | Palte, R.L. | | Deposit date: | 2020-10-30 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structure-Based Discovery of Proline-Derived Arginase Inhibitors with Improved Oral Bioavailability for Immuno-Oncology.
Acs Med.Chem.Lett., 12, 2021
|
|
7KLL
 
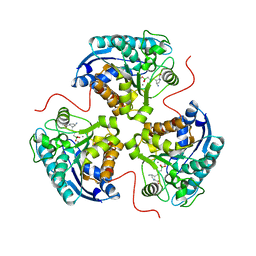 | | Human Arginase1 Complexed with Inhibitor Compound 18 | | Descriptor: | 3-[(2~{S},3~{R},4~{R})-4-azanyl-2-carboxy-pyrrolidin-3-yl]propyl-$l^{3}-oxidanyl-bis(oxidanyl)boron, Arginase-1, MANGANESE (II) ION | | Authors: | Palte, R.L. | | Deposit date: | 2020-10-30 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structure-Based Discovery of Proline-Derived Arginase Inhibitors with Improved Oral Bioavailability for Immuno-Oncology.
Acs Med.Chem.Lett., 12, 2021
|
|
7KLK
 
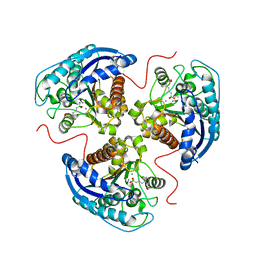 | | Human Arginase1 Complexed with Inhibitor Compound 3a | | Descriptor: | 1,2-ETHANEDIOL, 3-[(2~{S},3~{R})-2-carboxypiperidin-3-yl]propyl-$l^{3}-oxidanyl-bis(oxidanyl)boranuide, Arginase-1, ... | | Authors: | Palte, R.L. | | Deposit date: | 2020-10-30 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structure-Based Discovery of Proline-Derived Arginase Inhibitors with Improved Oral Bioavailability for Immuno-Oncology.
Acs Med.Chem.Lett., 12, 2021
|
|
7LWJ
 
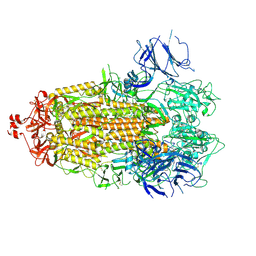 | |
7LWI
 
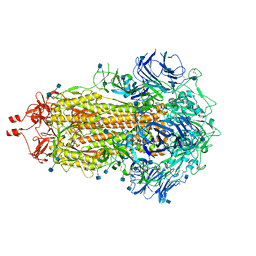 | |
7LWM
 
 | |
7LWW
 
 | |
7LWO
 
 | |
7LYM
 
 | |
7LWT
 
 | |
7LWU
 
 | |
7LWV
 
 | |
7LWK
 
 | |
7LWQ
 
 | |
7LWL
 
 | |
7LWP
 
 | |
7LWN
 
 | |
7KPY
 
 | | Crystal structure of CBP bromodomain liganded with UMB298 (compound 23) | | Descriptor: | 1,2-ETHANEDIOL, 2-[2-(3-chloranyl-4-methoxy-phenyl)ethyl]-~{N}-cyclohexyl-7-(3,5-dimethyl-1,2-oxazol-4-yl)imidazo[1,2-a]pyridin-3-amine, Histone acetyltransferase | | Authors: | Schonbrunn, E, Bikowitz, M. | | Deposit date: | 2020-11-12 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Development of Dimethylisoxazole-Attached Imidazo[1,2- a ]pyridines as Potent and Selective CBP/P300 Inhibitors.
J.Med.Chem., 64, 2021
|
|
8F9K
 
 | |
5YMU
 
 | |
5YLS
 
 | | Crystal structure of T2R-TTL-Y50 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, E-3-(3-azanyl-4-methoxy-phenyl)-1-(5-methoxy-2,2-dimethyl-chromen-8-yl)prop-2-en-1-one, ... | | Authors: | Yang, J.H, Chen, L.J. | | Deposit date: | 2017-10-18 | | Release date: | 2018-04-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The compound millepachine and its derivatives inhibit tubulin polymerization by irreversibly binding to the colchicine-binding site in beta-tubulin.
J. Biol. Chem., 293, 2018
|
|
5YLJ
 
 | | Crystal structure of T2R-TTL-Millepachine complex | | Descriptor: | (E)-1-(5-methoxy-2,2-dimethyl-chromen-8-yl)-3-(4-methoxyphenyl)prop-2-en-1-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Yang, J.H, Chen, L.J. | | Deposit date: | 2017-10-17 | | Release date: | 2018-04-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The compound millepachine and its derivatives inhibit tubulin polymerization by irreversibly binding to the colchicine-binding site in beta-tubulin.
J. Biol. Chem., 293, 2018
|
|
5YMT
 
 | |
5YMS
 
 | |
8H03
 
 | |
