8W31
 
 | | Crystal structure of parkin (R0RB):2pUb with activator compound | | Descriptor: | (S)-1-(6-benzyl-3-(4-(1,2,3,4-tetrahydroquinoline-1-carbonyl)phenyl)-6,7-dihydropyrazolo[1,5-a]pyrazin-5(4H)-yl)ethan-1-one, E3 ubiquitin-protein ligase parkin, GLYCEROL, ... | | Authors: | Sauve, V, Gehring, K. | | Deposit date: | 2024-02-21 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Activation of parkin by a molecular glue.
Nat Commun, 15, 2024
|
|
7NG2
 
 | | Crystal structure of Toxoplasma CPSF4-YTH domain in apo form | | Descriptor: | ISOPROPYL ALCOHOL, Zinc finger (CCCH type) motif-containing protein | | Authors: | Swale, C, Bowler, M.W. | | Deposit date: | 2021-02-08 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | A plant-like mechanism coupling m6A reading to polyadenylation safeguards transcriptome integrity and developmental gene partitioning in Toxoplasma .
Elife, 10, 2021
|
|
7NJC
 
 | |
7NH2
 
 | | Crystal structure of Toxoplasma CPSF4-YTH domain bound to m6A | | Descriptor: | TETRAETHYLENE GLYCOL, Zinc finger (CCCH type) motif-containing protein, ~{N},9-dimethylpurin-6-amine | | Authors: | Swale, C, Bowler, M.W. | | Deposit date: | 2021-02-09 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A plant-like mechanism coupling m6A reading to polyadenylation safeguards transcriptome integrity and developmental gene partitioning in Toxoplasma .
Elife, 10, 2021
|
|
7PIK
 
 | | Cryo-EM structure of E. coli TnsB in complex with right end fragment of Tn7 transposon | | Descriptor: | Right end fragment of Tn7 transposon, Transposon Tn7 transposition protein TnsB | | Authors: | Kaczmarska, Z, Czarnocki-Cieciura, M, Rawski, M, Nowotny, M. | | Deposit date: | 2021-08-20 | | Release date: | 2022-06-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Structural basis of transposon end recognition explains central features of Tn7 transposition systems.
Mol.Cell, 82, 2022
|
|
6BN1
 
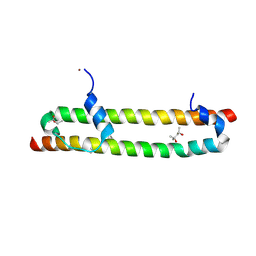 | | Salvador Hippo SARAH domain complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, NICKEL (II) ION, Scaffold protein salvador, ... | | Authors: | Cairns, L, Kavran, J.M. | | Deposit date: | 2017-11-15 | | Release date: | 2018-03-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Salvador has an extended SARAH domain that mediates binding to Hippo kinase.
J. Biol. Chem., 293, 2018
|
|
6IGX
 
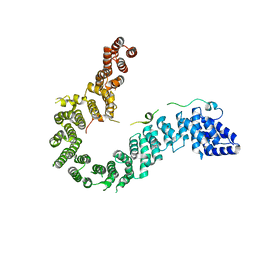 | | Crystal structure of human CAP-G in complex with CAP-H | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Condensin complex subunit 2, Condensin complex subunit 3 | | Authors: | Hara, K, Migita, T, Shimizu, K, Hashimoto, H. | | Deposit date: | 2018-09-26 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.995 Å) | | Cite: | Structural basis of HEAT-kleisin interactions in the human condensin I subcomplex.
Embo Rep., 20, 2019
|
|
8C1V
 
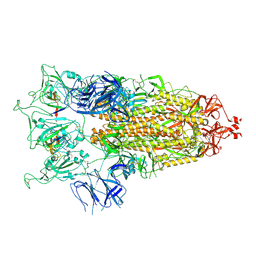 | | SARS-CoV-2 S-trimer (3 RBDs up) bound to TriSb92, fitted into cryo-EM map | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Sb92, ... | | Authors: | Huiskonen, J.T, Rissanen, I, Hannula, L. | | Deposit date: | 2022-12-21 | | Release date: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Intranasal trimeric sherpabody inhibits SARS-CoV-2 including recent immunoevasive Omicron subvariants.
Nat Commun, 14, 2023
|
|
7ZL3
 
 | |
3F2M
 
 | | Urate oxidase complexed with 8-azaxanthine at 150 MPa | | Descriptor: | 8-AZAXANTHINE, SODIUM ION, Uricase | | Authors: | Colloc'h, N, Girard, E, Kahn, R, Fourme, R. | | Deposit date: | 2008-10-30 | | Release date: | 2009-11-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-function perturbation and dissociation of tetrameric urate oxidase by high hydrostatic pressure.
Biophys.J., 98, 2010
|
|
4OYS
 
 | | CRYSTAL STRUCTURE OF VPS34 IN COMPLEX WITH SAR405. | | Descriptor: | (8S)-9-[(5-chloranylpyridin-3-yl)methyl]-2-[(3R)-3-methylmorpholin-4-yl]-8-(trifluoromethyl)-6,7,8,9a-tetrahydro-3H-pyrimido[1,2-a]pyrimidin-4-one, Phosphatidylinositol 3-kinase catalytic subunit type 3, SULFATE ION | | Authors: | Mathieu, M, Marquette, J.p. | | Deposit date: | 2014-02-13 | | Release date: | 2014-10-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A highly potent and selective Vps34 inhibitor alters vesicle trafficking and autophagy.
Nat.Chem.Biol., 10, 2014
|
|
2Z23
 
 | | Crystal structure of Y.pestis oligo peptide binding protein OppA with tri-lysine ligand | | Descriptor: | Periplasmic oligopeptide-binding protein, peptide (LYS)(LYS)(LYS) | | Authors: | Tanabe, M, Bertland, T, Mirza, O, Byrne, B, Brown, K.A. | | Deposit date: | 2007-05-17 | | Release date: | 2007-10-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of OppA and PstS from Yersinia pestis indicate variability of interactions with transmembrane domains.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
6PNW
 
 | |
6SAI
 
 | |
1B2K
 
 | | Structural effects of monovalent anions on polymorphic lysozyme crystals | | Descriptor: | IODIDE ION, PROTEIN (LYSOZYME) | | Authors: | Vaney, M.C, Broutin, I, Ries-Kautt, M, Ducruix, A. | | Deposit date: | 1998-11-26 | | Release date: | 1998-12-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural effects of monovalent anions on polymorphic lysozyme crystals.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
5H7U
 
 | | NMR structure of eIF3 36-163 | | Descriptor: | Eukaryotic translation initiation factor 3 subunit C | | Authors: | Nagata, T, Obayashi, E. | | Deposit date: | 2016-11-21 | | Release date: | 2017-05-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Molecular Landscape of the Ribosome Pre-initiation Complex during mRNA Scanning: Structural Role for eIF3c and Its Control by eIF5.
Cell Rep, 18, 2017
|
|
6SSL
 
 | |
6SA9
 
 | |
1MSC
 
 | |
6I2U
 
 | |
6I40
 
 | | Crystal structure of murine neuroglobin bound to CO at 15K under illumination using optical fiber | | Descriptor: | ACETATE ION, CARBON MONOXIDE, FORMIC ACID, ... | | Authors: | Savino, C, Montemiglio, L.C, Ardiccioni, C, Exertier, C, Vallone, B. | | Deposit date: | 2018-11-08 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ligand pathways in neuroglobin revealed by low-temperature photodissociation and docking experiments.
Iucrj, 6, 2019
|
|
5E95
 
 | | Crystal Structure of Mb(NS1)/H-Ras Complex | | Descriptor: | GTPase HRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Eguchi, R.R, Sha, F, Gupta, A, Koide, A, Koide, S. | | Deposit date: | 2015-10-14 | | Release date: | 2016-11-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.402 Å) | | Cite: | Inhibition of RAS function through targeting an allosteric regulatory site.
Nat. Chem. Biol., 13, 2017
|
|
6SSJ
 
 | |
2RVH
 
 | | NMR structure of eIF1 | | Descriptor: | Eukaryotic translation initiation factor eIF-1 | | Authors: | Nagata, T, Obayashi, E, Asano, K. | | Deposit date: | 2015-10-16 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Molecular Landscape of the Ribosome Pre-initiation Complex during mRNA Scanning: Structural Role for eIF3c and Its Control by eIF5
Cell Rep, 18, 2017
|
|
6SSM
 
 | |
