7RE7
 
 | | TCR mimic antibody (Fab fragment) in complex with AFP/HLA-A*02 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-2-microglobulin, FORMIC ACID, ... | | Authors: | Dasgupta, M, Baker, B.M. | | Deposit date: | 2021-07-12 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.547 Å) | | Cite: | Validation and promise of a TCR mimic antibody for cancer immunotherapy of hepatocellular carcinoma.
Sci Rep, 12, 2022
|
|
6EFJ
 
 | | Crystal structure of NDM-1 with compound 9 | | Descriptor: | (2R)-2-phenyl-2-(phenylamino)-N-(1H-tetrazol-5-yl)acetamide, Metallo-beta-lactamase type 2, ZINC ION | | Authors: | Akhtar, A, Chen, Y. | | Deposit date: | 2018-08-16 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Active-Site Druggability of Carbapenemases and Broad-Spectrum Inhibitor Discovery.
Acs Infect Dis., 5, 2019
|
|
7SK4
 
 | | Cryo-EM structure of ACKR3 in complex with chemokine N-terminal mutant CXCL12_LRHQ, an intracellular Fab, and an extracellular Fab | | Descriptor: | Atypical chemokine receptor 3, CHOLESTEROL, CID24 Fab heavy chain, ... | | Authors: | Yen, Y.C, Schafer, C.T, Gustavsson, M, Handel, T.M, Tesmer, J.J.G. | | Deposit date: | 2021-10-19 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of atypical chemokine receptor 3 reveal the basis for its promiscuity and signaling bias.
Sci Adv, 8, 2022
|
|
7SK6
 
 | | Cryo-EM structure of human ACKR3 in complex with chemokine N-terminal mutant CXCL12_LRHQ and an intracellular Fab | | Descriptor: | Atypical chemokine receptor 3, CID24 Fab heavy chain, CID24 Fab light chain, ... | | Authors: | Yen, Y.C, Schafer, C.T, Gustavsson, M, Handel, T.M, Tesmer, J.J.G. | | Deposit date: | 2021-10-19 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures of atypical chemokine receptor 3 reveal the basis for its promiscuity and signaling bias.
Sci Adv, 8, 2022
|
|
7SK9
 
 | | Cryo-EM structure of human ACKR3 in complex with a small molecule partial agonist CCX662, and an intracellular Fab | | Descriptor: | (1R)-4-[7-(3-carboxypropoxy)-6-methylquinolin-8-yl]-1-{[2-(4-hydroxypiperidin-1-yl)-1,3-thiazol-4-yl]methyl}-1,4-diazepan-1-ium, Atypical chemokine receptor 3, CHOLESTEROL, ... | | Authors: | Yen, Y.C, Schafer, C.T, Gustavsson, M, Handel, T.M, Tesmer, J.J.G. | | Deposit date: | 2021-10-19 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structures of atypical chemokine receptor 3 reveal the basis for its promiscuity and signaling bias.
Sci Adv, 8, 2022
|
|
7SK3
 
 | | Cryo-EM structure of ACKR3 in complex with CXCL12, an intracellular Fab, and an extracellular Fab | | Descriptor: | Atypical chemokine receptor 3, CHOLESTEROL, CID24 Fab heavy chain, ... | | Authors: | Yen, Y.C, Schafer, C.T, Gustavsson, M, Handel, T.M, Tesmer, J.J.G. | | Deposit date: | 2021-10-19 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of atypical chemokine receptor 3 reveal the basis for its promiscuity and signaling bias.
Sci Adv, 8, 2022
|
|
7SK8
 
 | | Cryo-EM structure of human ACKR3 in complex with CXCL12, a small molecule partial agonist CCX662, an extracellular Fab, and an intracellular Fab | | Descriptor: | (1R)-4-[7-(3-carboxypropoxy)-6-methylquinolin-8-yl]-1-{[2-(4-hydroxypiperidin-1-yl)-1,3-thiazol-4-yl]methyl}-1,4-diazepan-1-ium, Atypical chemokine receptor 3, CHOLESTEROL, ... | | Authors: | Yen, Y.C, Schafer, C.T, Gustavsson, M, Handel, T.M, Tesmer, J.J.G. | | Deposit date: | 2021-10-19 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of atypical chemokine receptor 3 reveal the basis for its promiscuity and signaling bias.
Sci Adv, 8, 2022
|
|
7SK5
 
 | | Cryo-EM structure of ACKR3 in complex with CXCL12 and an intracellular Fab | | Descriptor: | Anti-Fab nanobody, Atypical chemokine receptor 3, CHOLESTEROL, ... | | Authors: | Yen, Y.C, Schafer, C.T, Gustavsson, M, Handel, T.M, Tesmer, J.J.G. | | Deposit date: | 2021-10-19 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures of atypical chemokine receptor 3 reveal the basis for its promiscuity and signaling bias.
Sci Adv, 8, 2022
|
|
7SK7
 
 | | Cryo-EM structure of human ACKR3 in complex with CXCL12, a small molecule partial agonist CCX662, and an extracellular Fab | | Descriptor: | (1R)-4-[7-(3-carboxypropoxy)-6-methylquinolin-8-yl]-1-{[2-(4-hydroxypiperidin-1-yl)-1,3-thiazol-4-yl]methyl}-1,4-diazepan-1-ium, Anti-Fab nanobody, Atypical chemokine receptor 3, ... | | Authors: | Yen, Y.C, Schafer, C.T, Gustavsson, M, Handel, T.M, Tesmer, J.J.G. | | Deposit date: | 2021-10-19 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of atypical chemokine receptor 3 reveal the basis for its promiscuity and signaling bias.
Sci Adv, 8, 2022
|
|
6MHR
 
 | | Structure of the human 4-1BB / Urelumab Fab complex | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, MALONATE ION, ... | | Authors: | Kimberlin, C.R, Chin, S.M, Roe-Zurz, Z, Xu, A, Yang, Y. | | Deposit date: | 2018-09-18 | | Release date: | 2018-11-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the 4-1BB/4-1BBL complex and distinct binding and functional properties of utomilumab and urelumab.
Nat Commun, 9, 2018
|
|
6MI2
 
 | | Structure of the human 4-1BB / Utomilumab Fab complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, SULFATE ION, ... | | Authors: | Kimberlin, C.R, Chin, S.M, Roe-Zurz, Z, Xu, A, Yang, Y. | | Deposit date: | 2018-09-19 | | Release date: | 2018-11-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structure of the 4-1BB/4-1BBL complex and distinct binding and functional properties of utomilumab and urelumab.
Nat Commun, 9, 2018
|
|
6MGP
 
 | | Structure of human 4-1BB / 4-1BBL complex | | Descriptor: | ACETATE ION, GLYCEROL, Tumor necrosis factor ligand superfamily member 9, ... | | Authors: | Kimberlin, C.R, Chin, S.M, Roe-Zurz, Z, Xu, A, Yang, Y. | | Deposit date: | 2018-09-14 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structure of the 4-1BB/4-1BBL complex and distinct binding and functional properties of utomilumab and urelumab
Nat Commun, 9, 2018
|
|
5UQV
 
 | |
6MGE
 
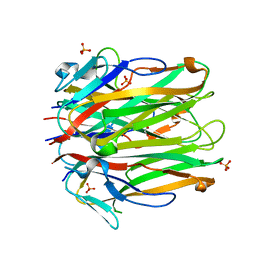 | | Structure of human 4-1BBL | | Descriptor: | GLYCEROL, PHOSPHATE ION, Tumor necrosis factor ligand superfamily member 9 | | Authors: | Kimberlin, C.R, Chin, S.M, Roe-Zurz, Z, Xu, A, Yang, Y. | | Deposit date: | 2018-09-13 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of the 4-1BB/4-1BBL complex and distinct binding and functional properties of utomilumab and urelumab.
Nat Commun, 9, 2018
|
|
5UQX
 
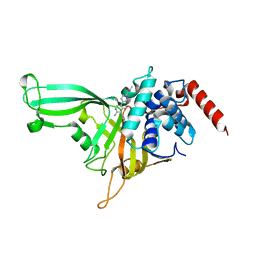 | |
6VBP
 
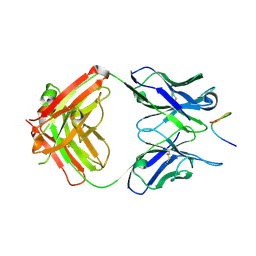 | |
6VBQ
 
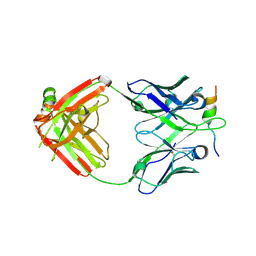 | |
6VBO
 
 | |
3BXV
 
 | |
6MD8
 
 | | Crystal structure of CTX-M-14 with compound 3 | | Descriptor: | 1-(2,4-dichlorophenyl)-4-(1H-tetrazol-5-yl)-1H-pyrazol-5-amine, Beta-lactamase CTX-M-14, DIMETHYL SULFOXIDE | | Authors: | Akhtar, A, Chen, Y. | | Deposit date: | 2018-09-04 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Active-Site Druggability of Carbapenemases and Broad-Spectrum Inhibitor Discovery.
Acs Infect Dis., 5, 2019
|
|
6MEY
 
 | | Crystal structure of KPC-2 with compound 9 | | Descriptor: | (2R)-2-phenyl-2-(phenylamino)-N-(1H-tetrazol-5-yl)acetamide, ACETATE ION, Carbapenem-hydrolyzing beta-lactamase KPC | | Authors: | Akhtar, A, Chen, Y. | | Deposit date: | 2018-09-07 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Active-Site Druggability of Carbapenemases and Broad-Spectrum Inhibitor Discovery.
Acs Infect Dis., 5, 2019
|
|
4XO7
 
 | | Crystal structure of human 3-alpha hydroxysteroid dehydrogenase type 3 in complex with NADP+, 5alpha-androstan-3,17-dione and (3beta, 5alpha)-3-hydroxyandrostan-17-one | | Descriptor: | 4-ANDROSTENE-3-17-DIONE, Aldo-keto reductase family 1 member C2, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Zhang, B, Hu, X.-J, Lin, S.-X. | | Deposit date: | 2015-01-16 | | Release date: | 2016-02-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Human 3 alpha-hydroxysteroid dehydrogenase type 3: structural clues of 5 alpha-DHT reverse binding and enzyme down-regulation decreasing MCF7 cell growth.
Biochem.J., 473, 2016
|
|
8QJR
 
 | | BRG1 bromodomain in complex with VBC via compound 17 | | Descriptor: | (2S,4R)-1-[(2R)-2-[3-[2-[4-[3-[4-[(1R,5S)-3-[3-azanyl-6-(2-hydroxyphenyl)pyridazin-4-yl]-3,8-diazabicyclo[3.2.1]octan-8-yl]pyridin-2-yl]oxycyclobutyl]oxypiperidin-1-yl]ethoxy]-1,2-oxazol-5-yl]-3-methyl-butanoyl]-N-[(1S)-1-[4-(4-methyl-1,3-thiazol-5-yl)phenyl]ethyl]-4-oxidanyl-pyrrolidine-2-carboxamide, CHLORIDE ION, Elongin-B, ... | | Authors: | Kerry, P.S, Hole, A.J, Perez-Dorado, J.I. | | Deposit date: | 2023-09-13 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | PROTACs Targeting BRM (SMARCA2) Afford Selective In Vivo Degradation over BRG1 (SMARCA4) and Are Active in BRG1 Mutant Xenograft Tumor Models.
J.Med.Chem., 67, 2024
|
|
6M7I
 
 | | Crystal structure of KPC-2 with compound 3 | | Descriptor: | 1-(2,4-dichlorophenyl)-4-(1H-tetrazol-5-yl)-1H-pyrazol-5-amine, Carbapenem-hydrolyzing beta-lactamase KPC, GLYCEROL | | Authors: | Akhtar, A, Chen, Y. | | Deposit date: | 2018-08-20 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Active-Site Druggability of Carbapenemases and Broad-Spectrum Inhibitor Discovery.
Acs Infect Dis., 5, 2019
|
|
6MIA
 
 | | Crystal structure of CTX-M-14 with compound 6 | | Descriptor: | 3-(1H-tetrazol-5-ylmethyl)-5,6,7,8-tetrahydro[1]benzothieno[2,3-d]pyrimidin-4(3H)-one, Beta-lactamase | | Authors: | Akhtar, A, Chen, Y. | | Deposit date: | 2018-09-19 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Active-Site Druggability of Carbapenemases and Broad-Spectrum Inhibitor Discovery.
Acs Infect Dis., 5, 2019
|
|
