7WLZ
 
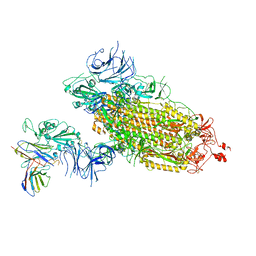 | |
2OJU
 
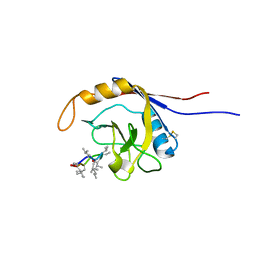 | | X-ray structure of complex of human cyclophilin J with cyclosporin A | | Descriptor: | CYCLOSPORIN A, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE-LIKE 3 | | Authors: | Xia, Z, Huang, L. | | Deposit date: | 2007-01-14 | | Release date: | 2008-01-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Targeting Cyclophilin J, a Novel Peptidyl-Prolyl Isomerase, Can Induce Cellular G1/S Arrest and Repress the Growth of Hepatocellular Carcinoma
To be Published
|
|
2OK3
 
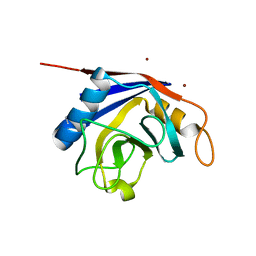 | | X-ray structure of human cyclophilin J at 2.0 angstrom | | Descriptor: | NICKEL (II) ION, Peptidyl-prolyl cis-trans isomerase-like 3 | | Authors: | Xia, Z. | | Deposit date: | 2007-01-15 | | Release date: | 2008-01-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Targeting Cyclophilin J, a novel peptidyl-prolyl isomerase, can induce cellular G1/S arrest and repress the growth of Hepatocellular carcinoma
To be Published
|
|
4PW2
 
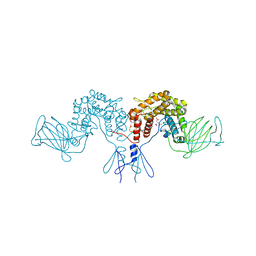 | | Crystal structure of D-glucuronyl C5 epimerase | | Descriptor: | CITRIC ACID, D-glucuronyl C5 epimerase B | | Authors: | Ke, J, Qin, Y, Gu, X, Brunzelle, J.S, Xu, H.E, Ding, K. | | Deposit date: | 2014-03-18 | | Release date: | 2015-01-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Functional Study of d-Glucuronyl C5-epimerase.
J.Biol.Chem., 290, 2015
|
|
5ZF2
 
 | | Crystal structure of Trxlp from Edwardsiella tarda EIB202 | | Descriptor: | SULFATE ION, Thioredoxin (H-type,TRX-H) | | Authors: | Yang, C, Quan, S. | | Deposit date: | 2018-03-02 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The Edwardsiella piscicida thioredoxin-like protein inhibits ASK1-MAPKs signaling cascades to promote pathogenesis during infection.
Plos Pathog., 15, 2019
|
|
6K0B
 
 | | cryo-EM structure of archaeal Ribonuclease P with mature tRNA | | Descriptor: | 50S ribosomal protein L7Ae, RPR, Ribonuclease P protein component 1, ... | | Authors: | Wan, F, Lan, P, Wu, J, Lei, M. | | Deposit date: | 2019-05-05 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-electron microscopy structure of an archaeal ribonuclease P holoenzyme.
Nat Commun, 10, 2019
|
|
6K0A
 
 | | cryo-EM structure of an archaeal Ribonuclease P | | Descriptor: | 50S ribosomal protein L7Ae, RPR, Ribonuclease P protein component 1, ... | | Authors: | Wan, F, Lan, P, Wu, J, Lei, M. | | Deposit date: | 2019-05-05 | | Release date: | 2019-06-19 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Cryo-electron microscopy structure of an archaeal ribonuclease P holoenzyme.
Nat Commun, 10, 2019
|
|
5A9Y
 
 | | Structure of ppGpp BipA | | Descriptor: | GTP-BINDING PROTEIN, GUANOSINE-5',3'-TETRAPHOSPHATE | | Authors: | Kumar, V, Chen, Y, Ero, R, Li, Z, Gao, Y.-G. | | Deposit date: | 2015-07-23 | | Release date: | 2015-08-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structure of Bipa in GTP Form Bound to the Ratcheted Ribosome.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5A9X
 
 | | Structure of GDP bound BipA | | Descriptor: | GTP-BINDING PROTEIN, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Kumar, V, Chen, Y, Ero, R, Li, Z, Gao, Y.-G. | | Deposit date: | 2015-07-23 | | Release date: | 2015-08-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure of Bipa in GTP Form Bound to the Ratcheted Ribosome.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5A9W
 
 | | Structure of GDPCP BipA | | Descriptor: | GTP-BINDING PROTEIN, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER | | Authors: | Kumar, V, Chen, Y, Ero, R, Li, Z, Gao, Y. | | Deposit date: | 2015-07-23 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structure of Bipa in GTP Form Bound to the Ratcheted Ribosome.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5A9V
 
 | | Structure of apo BipA | | Descriptor: | GTP-BINDING PROTEIN | | Authors: | Kumar, V, Chen, Y, Ero, R, Li, Z, Gao, Y. | | Deposit date: | 2015-07-23 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Structure of Bipa in GTP Form Bound to the Ratcheted Ribosome.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7OC7
 
 | | LasB, alpha-alkyl-N-aryl mercaptoacetamide | | Descriptor: | (2R)-N,3-diphenyl-2-sulfanyl-propanamide, CALCIUM ION, Neutral metalloproteinase, ... | | Authors: | Koehnke, J, Sikandar, A. | | Deposit date: | 2021-04-26 | | Release date: | 2022-01-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Substrate-Inspired Fragment Merging and Growing Affords Efficacious LasB Inhibitors.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
6IUY
 
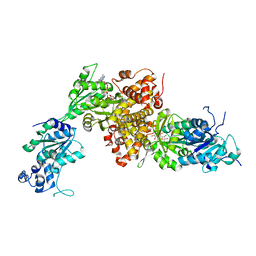 | | Structure of DsGPDH of Dunaliella salina | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, GLYCEROL, Glycerol-3-phosphate dehydrogenase [NAD(+)], ... | | Authors: | He, Q, Toh, J.D, Ero, R, Qiao, Z, Kumar, V, Gao, Y.G. | | Deposit date: | 2018-12-01 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The unusual di-domain structure of Dunaliella salina glycerol-3-phosphate dehydrogenase enables direct conversion of dihydroxyacetone phosphate to glycerol.
Plant J., 102, 2020
|
|
6K2U
 
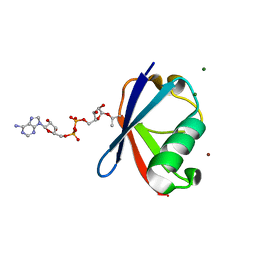 | | Crystal structure of Thr66 ADP-ribosylated ubiquitin | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, MAGNESIUM ION, Polyubiquitin-C, ... | | Authors: | Wang, X, Zhou, Y, Zhu, Y. | | Deposit date: | 2019-05-15 | | Release date: | 2020-03-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.554 Å) | | Cite: | Threonine ADP-Ribosylation of Ubiquitin by a Bacterial Effector Family Blocks Host Ubiquitination.
Mol.Cell, 78, 2020
|
|
9JS4
 
 | | Cryo-EM structure of neutralizing antibody 8G3 in complex with BA.1 RBD | | Descriptor: | Heavy chain of 8G3, Light chain of 8G3, Spike glycoprotein | | Authors: | Li, J, Li, H. | | Deposit date: | 2024-09-30 | | Release date: | 2025-01-22 | | Last modified: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Rapid restoration of potent neutralization activity against the latest Omicron variant JN.1 via AI rational design and antibody engineering.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9KR5
 
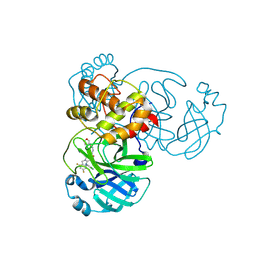 | | Crystal structure of SARS-CoV-2 main protease in complex with compound 3 | | Descriptor: | (6~{E})-6-(6-chloranyl-2-methyl-indazol-5-yl)imino-3-[5-[(3~{S})-oxolan-3-yl]oxypyridin-3-yl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazinane-2,4-dione, 3C-like proteinase nsp5 | | Authors: | Zhong, Y, Zhao, L, Zhang, W, Peng, W. | | Deposit date: | 2024-11-27 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Expanding the utilization of binding pockets proves to be effective for noncovalent small molecule inhibitors against SARS-CoV-2 M pro.
Eur.J.Med.Chem., 289, 2025
|
|
9KSK
 
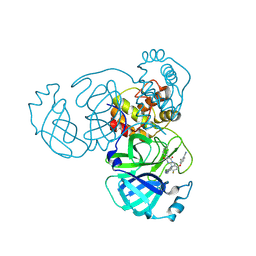 | | Crystal structure of SARS-CoV-2 main protease in complex with compound 10 | | Descriptor: | 3C-like proteinase, 4-[4-chloranyl-2-[[(6E)-6-(6-chloranyl-2-methyl-indazol-5-yl)imino-3-(5-methylpyridin-3-yl)-2,4-bis(oxidanylidene)-1,3,5-triazinan-1-yl]methyl]-5-fluoranyl-phenoxy]-2-fluoranyl-benzenecarbonitrile | | Authors: | Zhong, Y, Zhao, L, Zhang, W, Peng, W. | | Deposit date: | 2024-11-29 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Expanding the utilization of binding pockets proves to be effective for noncovalent small molecule inhibitors against SARS-CoV-2 M pro.
Eur.J.Med.Chem., 289, 2025
|
|
9KSI
 
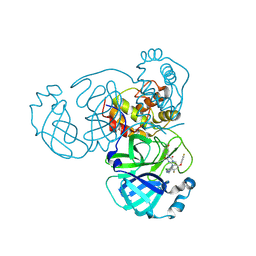 | | Crystal Structure of SARS-CoV-2 main protease in complex with compound 5 | | Descriptor: | (6E)-1-[[5-chloranyl-4-fluoranyl-2-(4-fluoranylphenoxy)phenyl]methyl]-6-(6-chloranyl-2-methyl-indazol-5-yl)imino-3-(5-methoxypyridin-3-yl)-1,3,5-triazinane-2,4-dione, 3C-like proteinase nsp5 | | Authors: | Zhong, Y, Zhao, L, Zhang, W, Peng, W. | | Deposit date: | 2024-11-29 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Expanding the utilization of binding pockets proves to be effective for noncovalent small molecule inhibitors against SARS-CoV-2 M pro.
Eur.J.Med.Chem., 289, 2025
|
|
9KSH
 
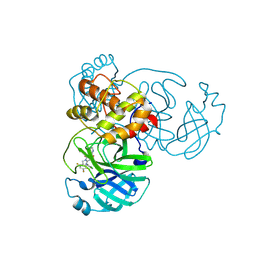 | | Crystal structure of SARS-CoV-2 main protease in complex with compound 1 | | Descriptor: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-pyridin-3-yl-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | Authors: | Zhong, Y, Zhao, L, Zhang, W, Peng, W. | | Deposit date: | 2024-11-29 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Expanding the utilization of binding pockets proves to be effective for noncovalent small molecule inhibitors against SARS-CoV-2 M pro.
Eur.J.Med.Chem., 289, 2025
|
|
9KSJ
 
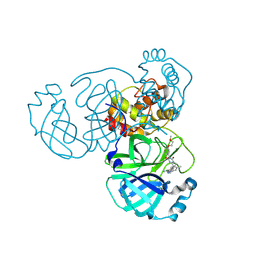 | | Crystal structure of SARS-CoV-2 main protease in complex with compound 8 | | Descriptor: | 3-[[(6E)-6-(6-chloranyl-2-methyl-indazol-5-yl)imino-3-[5-(2-methoxyethoxy)pyridin-3-yl]-2,4-bis(oxidanylidene)-1,3,5-triazinan-1-yl]methyl]-4-methyl-benzenecarbonitrile, 3C-like proteinase nsp5 | | Authors: | Zhong, Y, Zhao, L, Zhang, W, Peng, W. | | Deposit date: | 2024-11-29 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Expanding the utilization of binding pockets proves to be effective for noncovalent small molecule inhibitors against SARS-CoV-2 M pro.
Eur.J.Med.Chem., 289, 2025
|
|
2DBT
 
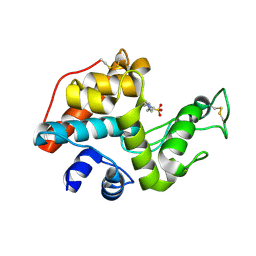 | |
5Z5S
 
 | | Crystal structure of the PPARgamma-LBD complexed with compound 13ab | | Descriptor: | 3-{[6-(4-chloro-3-fluorophenoxy)-1-methyl-1H-benzimidazol-2-yl]methoxy}benzoic acid, CHLORIDE ION, Peptide from Peroxisome proliferator-activated receptor gamma coactivator 1-alpha, ... | | Authors: | Matsui, Y, Hanzawa, H. | | Deposit date: | 2018-01-19 | | Release date: | 2018-10-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of DS-6930, a potent selective PPAR gamma modulator. Part I: Lead identification.
Bioorg. Med. Chem., 26, 2018
|
|
5Z6S
 
 | | Crystal structure of the PPARgamma-LBD complexed with compound DS-6930 | | Descriptor: | 3-[[6-(3,5-dimethylpyridin-2-yl)oxy-1-methyl-benzimidazol-2-yl]methoxy]benzoic acid, CHLORIDE ION, Peptide from Peroxisome proliferator-activated receptor gamma coactivator 1-alpha, ... | | Authors: | Matsui, Y, Hanzawa, H. | | Deposit date: | 2018-01-25 | | Release date: | 2018-10-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of DS-6930, a potent selective PPAR gamma modulator. Part II: Lead optimization.
Bioorg. Med. Chem., 26, 2018
|
|
8GOU
 
 | | Omicron BA.4/5 SARS-CoV-2 S in complex with TH003 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, TH003 Fab heavy chain, ... | | Authors: | Guo, Y, Zhang, G, Liang, J, Liu, F, Rao, Z. | | Deposit date: | 2022-08-25 | | Release date: | 2023-06-28 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
8GPY
 
 | | Crystal structure of Omicron BA.4/5 RBD in complex with a neutralizing antibody scFv | | Descriptor: | Spike protein S1, scFv | | Authors: | Gao, Y.X, Song, Z.D, Wang, W.M, Guo, Y. | | Deposit date: | 2022-08-27 | | Release date: | 2023-06-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
