3D91
 
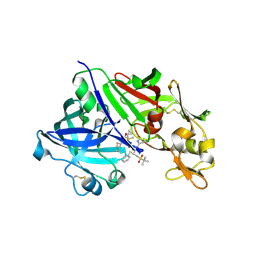 | | Human renin in complex with remikiren | | Descriptor: | DIMETHYL SULFOXIDE, Nalpha-[(2S)-2-benzyl-3-(tert-butylsulfonyl)propanoyl]-N-[(1S,2R,3S)-1-(cyclohexylmethyl)-3-cyclopropyl-2,3-dihydroxypropyl]-L-histidinamide, Renin | | Authors: | Prade, L, Bezencon, O, Bur, D, Weller, T, Fischli, W, Remen, L. | | Deposit date: | 2008-05-26 | | Release date: | 2008-06-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Human renin in complex with remikiren
to be published
|
|
3DFX
 
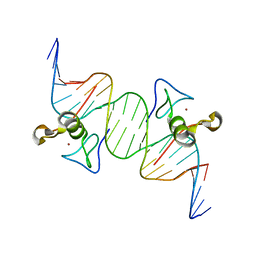 | | Opposite GATA DNA binding | | Descriptor: | DNA (5'-D(*DAP*DAP*DGP*DGP*DTP*DTP*DAP*DTP*DCP*DTP*DCP*DTP*DGP*DAP*DTP*DTP*DTP*DAP*DTP*DC)-3'), DNA (5'-D(*DTP*DTP*DGP*DAP*DTP*DAP*DAP*DAP*DTP*DCP*DAP*DGP*DAP*DGP*DAP*DTP*DAP*DAP*DCP*DC)-3'), Trans-acting T-cell-specific transcription factor GATA-3, ... | | Authors: | Bates, D.L, Kim, G.K, Guo, L, Chen, L. | | Deposit date: | 2008-06-12 | | Release date: | 2008-07-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of multiple GATA zinc fingers bound to DNA reveal new insights into DNA recognition and self-association by GATA.
J.Mol.Biol., 381, 2008
|
|
1M74
 
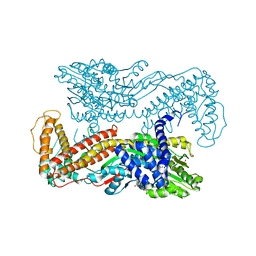 | | Crystal structure of Mg-ADP-bound SecA from Bacillus subtilis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Preprotein translocase secA, ... | | Authors: | Hunt, J.F, Weinkauf, S, Henry, L, Fak, J.J, McNicholas, P, Oliver, D.B, Deisenhofer, J. | | Deposit date: | 2002-07-16 | | Release date: | 2002-09-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Nucleotide Control of Interdomain Interactions in the Conformational Reaction Cycle of SecA
Science, 297, 2002
|
|
1MGR
 
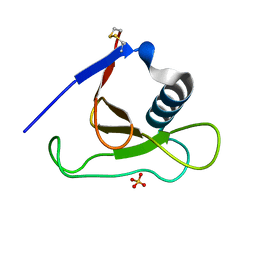 | | Crystal structure of RNase Sa3,cytotoxic microbial ribonuclease | | Descriptor: | Guanyl-specific ribonuclease Sa3, SULFATE ION | | Authors: | Sevcik, J, Urbanikova, L, Leland, P.A, Raines, R.T. | | Deposit date: | 2002-08-16 | | Release date: | 2003-02-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Links X-ray Structure of Two Crystalline Forms of a Streptomycete Ribonuclease with Cytotoxic Activity
J.Biol.Chem., 277, 2002
|
|
1MGW
 
 | | Crystal structure of RNase Sa3, cytotoxic microbial ribonuclease | | Descriptor: | Guanyl-specific ribonuclease Sa3, LITHIUM ION | | Authors: | Sevcik, J, Urbanikova, L, Leland, P.A, Raines, R.T. | | Deposit date: | 2002-08-16 | | Release date: | 2003-02-04 | | Last modified: | 2011-11-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Links X-ray Structure of Two Crystalline Forms of a Streptomycete Ribonuclease with Cytotoxic Activity
J.Biol.Chem., 277, 2002
|
|
3DH1
 
 | | Crystal structure of human tRNA-specific adenosine-34 deaminase subunit ADAT2 | | Descriptor: | ZINC ION, tRNA-specific adenosine deaminase 2 | | Authors: | Welin, M, Tresaugues, L, Andersson, J, Arrowsmith, C.H, Berglund, H, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kotenyova, T, Lehtio, L, Moche, M, Nilsson, M.E, Nyman, T, Olesen, K, Persson, C, Sagemark, J, Schueler, H, Thorsell, A.G, van der Berg, S, Wisniewska, M, Wikstrom, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-06-16 | | Release date: | 2008-09-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of human tRNA-specific adenosine-34 deaminase subunit ADAT2.
To be Published
|
|
3DOY
 
 | | Crystal structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase (FabZ) from Helicobacter pylori in complex with compound 3i | | Descriptor: | (3R)-hydroxymyristoyl-acyl carrier protein dehydratase, 4-chloro-N'-[(1E)-(3,5-dibromo-2,4-dihydroxyphenyl)methylidene]benzohydrazide, BENZAMIDINE, ... | | Authors: | Zhang, L, He, L, Liu, X, Liu, H, Shen, X, Jiang, H. | | Deposit date: | 2008-07-07 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovering potent inhibitors against the beta-hydroxyacyl-acyl carrier protein dehydratase (FabZ) of Helicobacter pylori: structure-based design, synthesis, bioassay, and crystal structure determination.
J.Med.Chem., 52, 2009
|
|
2H3A
 
 | | Structural basis for nucleic acid and toxin recognition of the bacterial antitoxin CcdA | | Descriptor: | 5'-D(P*AP*TP*AP*TP*GP*TP*AP*TP*AP*CP*CP*CP*G)-3', 5'-D(P*TP*CP*GP*GP*GP*TP*AP*TP*AP*CP*AP*TP*A)-3', CcdA | | Authors: | Madl, T, Van Melderen, L, Respondek, M, Oberer, M, Keller, W, Zangger, K. | | Deposit date: | 2006-05-22 | | Release date: | 2006-11-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Nucleic Acid and Toxin Recognition of the Bacterial Antitoxin CcdA
J.Mol.Biol., 364, 2006
|
|
2GLM
 
 | | Crystal structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase(FabZ) from Helicobacter pylori complexed with Compound 2 | | Descriptor: | (3R)-hydroxymyristoyl-acyl carrier protein dehydratase, 2-CHLORO-5-(5-{(E)-[(2Z)-3-(2-METHOXYETHYL)-4-OXO-2-(PHENYLIMINO)-1,3-THIAZOLIDIN-5-YLIDENE]METHYL}-2-FURYL)BENZOIC ACID, BENZAMIDINE, ... | | Authors: | Zhang, L, Liu, W, Shen, X, Jiang, H. | | Deposit date: | 2006-04-05 | | Release date: | 2007-03-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for catalytic and inhibitory mechanisms of beta-hydroxyacyl-acyl carrier protein dehydratase (FabZ).
J.Biol.Chem., 283, 2008
|
|
3H9W
 
 | | Crystal Structure of the N-terminal domain of Diguanylate cyclase with PAS/PAC sensor (Maqu_2914) from Marinobacter aquaeolei, Northeast Structural Genomics Consortium Target MqR66C | | Descriptor: | Diguanylate cyclase with PAS/PAC sensor | | Authors: | Seetharaman, J, Su, M, Wang, H, Foote, E.L, Mao, L, Nair, R, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-04-30 | | Release date: | 2009-05-19 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Northeast Structural Genomics Consortium Target MqR66C
To be Published
|
|
1M4R
 
 | | CRYSTAL STRUCTURE OF RECOMBINANT HUMAN INTERLEUKIN-22 | | Descriptor: | Interleukin-22 | | Authors: | Nagem, R.A.P, Colau, D, Dumoutier, L, Renauld, J.-C, Ogata, C, Polikarpov, I. | | Deposit date: | 2002-07-03 | | Release date: | 2003-07-07 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Recombinant Human Interleukin-22
Structure, 10, 2002
|
|
2HP8
 
 | | SOLUTION STRUCTURE OF HUMAN P8-MTCP1, A CYSTEINE-RICH PROTEIN ENCODED BY THE MTCP1 ONCOGENE,REVEALS A NEW ALPHA-HELICAL ASSEMBLY MOTIF, NMR, 30 STRUCTURES | | Descriptor: | Cx9C motif-containing protein 4 | | Authors: | Barthe, P, Chiche, L, Strub, M.P, Roumestand, C. | | Deposit date: | 1997-08-26 | | Release date: | 1998-03-04 | | Last modified: | 2019-08-21 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human p8MTCP1, a cysteine-rich protein encoded by the MTCP1 oncogene, reveals a new alpha-helical assembly motif.
J.Mol.Biol., 274, 1997
|
|
1SXQ
 
 | |
3HIX
 
 | | Crystal Structure of the Rhodanese_3 like domain from Anabaena sp Alr3790 protein. Northeast Structural Genomics Consortium Target NsR437i | | Descriptor: | Alr3790 protein, MANGANESE (II) ION | | Authors: | Vorobiev, S, Chen, Y, Forouhar, F, Maglaqui, M, Ciccosanti, C, Mao, L, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-05-20 | | Release date: | 2009-05-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal Structure of the Rhodanese_3 like domain from Anabaena sp Alr3790 protein.
To be Published
|
|
1JPT
 
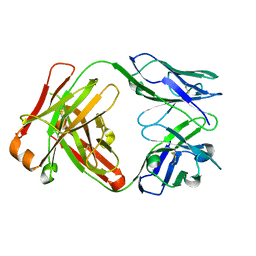 | | Crystal Structure of Fab D3H44 | | Descriptor: | immunoglobulin Fab D3H44, heavy chain, immunoglobulin Fab D3h44, ... | | Authors: | Faelber, K, Kirchhofer, D, Presta, L, Kelley, R.F, Muller, Y.A. | | Deposit date: | 2001-08-03 | | Release date: | 2002-02-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The 1.85 A resolution crystal structures of tissue factor in complex with humanized Fab D3h44 and of free humanized Fab D3h44: revisiting the solvation of antigen combining sites.
J.Mol.Biol., 313, 2001
|
|
3HFX
 
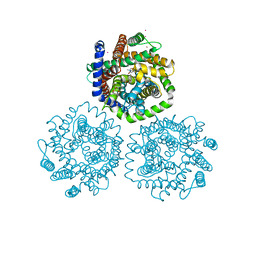 | | Crystal structure of carnitine transporter | | Descriptor: | CARNITINE, L-carnitine/gamma-butyrobetaine antiporter, MERCURY (II) ION | | Authors: | Tang, L, Wang, W.-H, Bai, L, Jiang, T. | | Deposit date: | 2009-05-13 | | Release date: | 2010-03-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Crystal structure of the carnitine transporter and insights into the antiport mechanism
Nat.Struct.Mol.Biol., 17, 2010
|
|
3HFQ
 
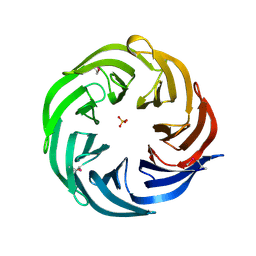 | | Crystal structure of the lp_2219 protein from Lactobacillus plantarum. Northeast Structural Genomics Consortium Target LpR118. | | Descriptor: | PHOSPHATE ION, uncharacterized protein lp_2219 | | Authors: | Vorobiev, S.M, Scott, L, Schauder, C, Xiao, R, Ciccosanti, C, Foote, E.L, Maglaqui, M, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-05-12 | | Release date: | 2009-05-26 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (1.963 Å) | | Cite: | Crystal structure of the lp_2219 protein from Lactobacillus plantarum.
To be Published
|
|
3HJB
 
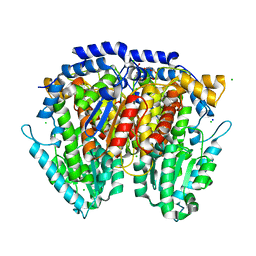 | | 1.5 Angstrom Crystal Structure of Glucose-6-phosphate Isomerase from Vibrio cholerae. | | Descriptor: | CALCIUM ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-05-21 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.5 Angstrom Crystal Structure of Glucose-6-phosphate Isomerase from Vibrio cholerae.
To be Published
|
|
1MEQ
 
 | | HIV gp120 C5 | | Descriptor: | Exterior Membrane Glycoprotein (GP120) | | Authors: | Caffrey, M, Jacobs, A, Guilhaudis, L. | | Deposit date: | 2002-08-08 | | Release date: | 2002-12-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the HIV gp120 C5 Domain
Eur.J.Biochem., 269, 2002
|
|
2JOO
 
 | | The NMR Solution Structure of Recombinant RGD-hirudin | | Descriptor: | Hirudin variant-1 | | Authors: | Song, X, Mo, W, Liu, X, Yan, X, Song, H, Dai, L. | | Deposit date: | 2007-03-14 | | Release date: | 2008-03-18 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structure of recombinant RGD-hirudin
Biochem.Biophys.Res.Commun., 360, 2007
|
|
1M5R
 
 | | Ternary complex of T4 phage BGT with UDP and a 13 mer DNA duplex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-D(*CP*TP*AP*TP*CP*TP*GP*AP*GP*TP*AP*TP*C)-3', ... | | Authors: | Lariviere, L, Morera, S. | | Deposit date: | 2002-07-10 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Base-flipping mechanism for the T4 phage beta-glucosyltransferase and
identification of a transition state analog
J.Mol.Biol., 324, 2002
|
|
1EE1
 
 | | CRYSTAL STRUCTURE OF NH3-DEPENDENT NAD+ SYNTHETASE FROM BACILLUS SUBTILIS COMPLEXED WITH ONE MOLECULE ATP, TWO MOLECULES DEAMIDO-NAD+ AND ONE MG2+ ION | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, NH(3)-DEPENDENT NAD(+) SYNTHETASE, ... | | Authors: | Devedjiev, Y, Symersky, J, Singh, R, Jedrzejas, M, Brouillette, C, Brouillette, W, Muccio, D, Chattopadhyay, D, Delucas, L. | | Deposit date: | 2000-01-28 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Stabilization of active-site loops in NH3-dependent NAD+ synthetase from Bacillus subtilis.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1M6N
 
 | | Crystal structure of the SecA translocation ATPase from Bacillus subtilis | | Descriptor: | Preprotein translocase secA, SULFATE ION | | Authors: | Hunt, J.F, Weinkauf, S, Henry, L, Fak, J.J, McNicholas, P, Oliver, D.B, Deisenhofer, J. | | Deposit date: | 2002-07-16 | | Release date: | 2002-09-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Nucleotide Control of Interdomain Interactions in the Conformational Reaction Cycle of SecA
Science, 297, 2002
|
|
1EI1
 
 | | DIMERIZATION OF E. COLI DNA GYRASE B PROVIDES A STRUCTURAL MECHANISM FOR ACTIVATING THE ATPASE CATALYTIC CENTER | | Descriptor: | DNA GYRASE B, GLYCEROL, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Brino, L, Urzhumtsev, A, Oudet, P, Moras, D. | | Deposit date: | 2000-02-23 | | Release date: | 2000-03-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Dimerization of Escherichia coli DNA-gyrase B provides a structural mechanism for activating the ATPase catalytic center.
J.Biol.Chem., 275, 2000
|
|
1VLX
 
 | | STRUCTURE OF ELECTRON TRANSFER (COBALT-PROTEIN) | | Descriptor: | AZURIN, COBALT (II) ION | | Authors: | Bonander, N, Vanngard, T, Tsai, L.-C, Langer, V, Nar, H, Sjolin, L. | | Deposit date: | 1996-10-08 | | Release date: | 1997-03-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The metal site of Pseudomonas aeruginosa azurin, revealed by a crystal structure determination of the Co(II) derivative and Co-EPR spectroscopy.
Proteins, 27, 1997
|
|
