5ZNT
 
 | | Insect chitin deacetylase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ZINC ION, ... | | Authors: | Liu, L, Zhou, Y, Yang, Q. | | Deposit date: | 2018-04-10 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.979 Å) | | Cite: | Structural and biochemical insights into the catalytic mechanisms of two insect chitin deacetylases of the carbohydrate esterase 4 family.
J. Biol. Chem., 294, 2019
|
|
7F0D
 
 | | Cryo-EM structure of Mycobacterium tuberculosis 50S ribosome subunit bound with clarithromycin | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Zhang, W, Sun, Y, Gao, N, Li, Z. | | Deposit date: | 2021-06-03 | | Release date: | 2022-06-29 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of Mycobacterium tuberculosis 50S ribosomal subunit bound with clarithromycin reveals dynamic and specific interactions with macrolides.
Emerg Microbes Infect, 11, 2022
|
|
6CPW
 
 | | Discovery of 3(S)-thiomethyl pyrrolidine ERK inhibitors for oncology | | Descriptor: | (3S)-N-[3-(4-fluorophenyl)-1H-indazol-5-yl]-3-(methylsulfanyl)-1-(2-oxo-2-{4-[4-(pyrimidin-2-yl)phenyl]piperazin-1-yl}ethyl)pyrrolidine-3-carboxamide, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Hruza, A, Hruza, A. | | Deposit date: | 2018-03-14 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of 3(S)-thiomethyl pyrrolidine ERK inhibitors for oncology.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
5ZNS
 
 | | Insect chitin deacetylase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ZINC ION, ... | | Authors: | Liu, L, Zhou, Y, Yang, Q. | | Deposit date: | 2018-04-10 | | Release date: | 2019-02-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.396 Å) | | Cite: | Structural and biochemical insights into the catalytic mechanisms of two insect chitin deacetylases of the carbohydrate esterase 4 family.
J. Biol. Chem., 294, 2019
|
|
4ZZH
 
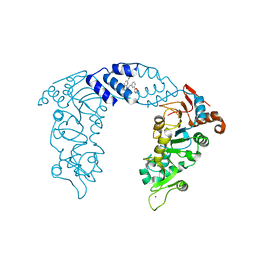 | | SIRT1/Activator Complex | | Descriptor: | (4S)-N-[3-(1,3-oxazol-5-yl)phenyl]-7-[3-(trifluoromethyl)phenyl]-3,4-dihydro-1,4-methanopyrido[2,3-b][1,4]diazepine-5(2H)-carboxamide, NAD-dependent protein deacetylase sirtuin-1, ZINC ION | | Authors: | Dai, H. | | Deposit date: | 2015-05-22 | | Release date: | 2015-07-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.1001 Å) | | Cite: | Crystallographic structure of a small molecule SIRT1 activator-enzyme complex.
Nat Commun, 6, 2015
|
|
7FEW
 
 | |
7FF0
 
 | |
4IVB
 
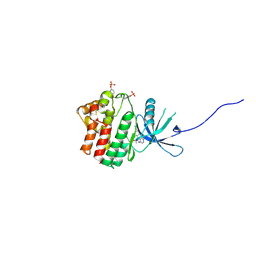 | | JAK1 kinase (JH1 domain) in complex with the inhibitor TRANS-4-{2-[(1R)-1-HYDROXYETHYL]IMIDAZO[4,5-D]PYRROLO[2,3-B]PYRIDIN-1(6H)-YL}CYCLOHEXANECARBONITRILE | | Descriptor: | Tyrosine-protein kinase JAK1, trans-4-{2-[(1R)-1-hydroxyethyl]imidazo[4,5-d]pyrrolo[2,3-b]pyridin-1(6H)-yl}cyclohexanecarbonitrile | | Authors: | Eigenbrot, C, Steffek, M. | | Deposit date: | 2013-01-22 | | Release date: | 2013-05-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of C-2 Hydroxyethyl Imidazopyrrolopyridines as Potent JAK1 Inhibitors with Favorable Physicochemical Properties and High Selectivity over JAK2.
J.Med.Chem., 56, 2013
|
|
7XPQ
 
 | |
3PZ8
 
 | |
7XPP
 
 | |
4IVC
 
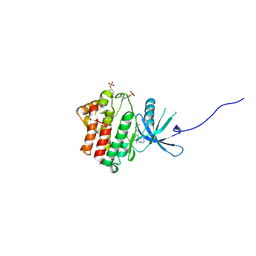 | | JAK1 kinase (JH1 domain) in complex with the inhibitor (TRANS-4-{2-[(1R)-1-HYDROXYETHYL]IMIDAZO[4,5-D]PYRROLO[2,3-B]PYRIDIN-1(6H)-YL}CYCLOHEXYL)ACETONITRILE | | Descriptor: | (trans-4-{2-[(1R)-1-hydroxyethyl]imidazo[4,5-d]pyrrolo[2,3-b]pyridin-1(6H)-yl}cyclohexyl)acetonitrile, Tyrosine-protein kinase JAK1 | | Authors: | Eigenbrot, C, Shia, S. | | Deposit date: | 2013-01-22 | | Release date: | 2013-05-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Identification of C-2 Hydroxyethyl Imidazopyrrolopyridines as Potent JAK1 Inhibitors with Favorable Physicochemical Properties and High Selectivity over JAK2.
J.Med.Chem., 56, 2013
|
|
4N6Z
 
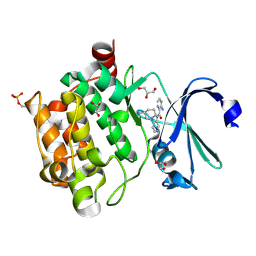 | | Pim1 Complexed with a pyridylcarboxamide | | Descriptor: | 3-amino-N-{4-[(3S)-3-aminopiperidin-1-yl]pyridin-3-yl}pyrazine-2-carboxamide, GLYCEROL, Serine/threonine-protein kinase pim-1 | | Authors: | Bellamacina, C.R, Le, V, Shu, W, Burger, M.T, Bussiere, D. | | Deposit date: | 2013-10-14 | | Release date: | 2013-11-06 | | Last modified: | 2014-07-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure Guided Optimization, in Vitro Activity, and in Vivo Activity of Pan-PIM Kinase Inhibitors.
ACS Med Chem Lett, 4, 2013
|
|
4YZD
 
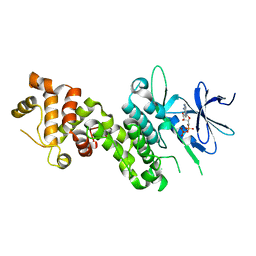 | | Crystal Structure of human phosphorylated IRE1alpha in complex with ADP-Mg | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Concha, N.O. | | Deposit date: | 2015-03-24 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | Long-Range Inhibitor-Induced Conformational Regulation of Human IRE1 alpha Endoribonuclease Activity.
Mol.Pharmacol., 88, 2015
|
|
4FS4
 
 | | Structure of BACE Bound to (S)-4-(3'-methoxy-[1,1'-biphenyl]-3-yl)-1,4-dimethyl-6-oxotetrahydropyrimidin-2(1H)-iminium | | Descriptor: | (6S)-2-amino-6-(3'-methoxybiphenyl-3-yl)-3,6-dimethyl-5,6-dihydropyrimidin-4(3H)-one, Beta-secretase 1, L(+)-TARTARIC ACID | | Authors: | Strickland, C, Stamford, A. | | Deposit date: | 2012-06-26 | | Release date: | 2012-10-10 | | Last modified: | 2014-07-23 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | A Potent and Orally Efficacious, Hydroxyethylamine-Based Inhibitor of beta-Secretase.
ACS Med Chem Lett, 3, 2012
|
|
5Z8Q
 
 | |
5Z8I
 
 | |
3HO9
 
 | | Structure of E.coli FabF(C163A) in complex with Platencin A1 | | Descriptor: | 2,4-dihydroxy-3-({3-[(2R,4aR,8S,8aR,9R)-9-hydroxy-8-methyl-3-methylidene-7-oxo-1,3,4,7,8,8a-hexahydro-2H-2,4a-ethanonap hthalen-8-yl]propanoyl}amino)benzoic acid, 3-oxoacyl-[acyl-carrier-protein] synthase 2 | | Authors: | Soisson, S.M, Parthasarathy, G. | | Deposit date: | 2009-06-01 | | Release date: | 2010-02-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Isolation, enzyme-bound structure and antibacterial activity of platencin A1 from Streptomyces platensis.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
8JV5
 
 | |
8JV6
 
 | |
3HNZ
 
 | |
5XLP
 
 | |
4KMA
 
 | |
6A85
 
 | | Crystal structure of a novel DNA quadruplex | | Descriptor: | AMMONIUM ION, DNA (5'-D(*AP*GP*AP*GP*AP*GP*AP*TP*GP*GP*GP*TP*GP*CP*GP*TP*T)-3'), LEAD (II) ION, ... | | Authors: | Liu, H.H, Gan, J.H. | | Deposit date: | 2018-07-06 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | High-resolution DNA quadruplex structure containing all the A-, G-, C-, T-tetrads.
Nucleic Acids Res., 46, 2018
|
|
7CH1
 
 | | The overall structure of SLC26A9 | | Descriptor: | CHLORIDE ION, SODIUM ION, Solute carrier family 26 member 9 | | Authors: | Chi, X.M, Chen, Y, Li, X.R, Zhang, Y.Y, Zhou, Q. | | Deposit date: | 2020-07-03 | | Release date: | 2020-08-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural insights into the gating mechanism of human SLC26A9 mediated by its C-terminal sequence.
Cell Discov, 6, 2020
|
|
