7CAH
 
 | | The interface of H014 Fab binds to SARS-CoV-2 S | | Descriptor: | Heavy chain of H014 Fab, Light chain of H014 Fab, Spike protein S1 | | Authors: | Zhe, L, Cao, L, Deng, Y, Sun, Y, Wang, N, Xie, L, Rao, Z, wang, Y, Qin, C, Wang, X. | | Deposit date: | 2020-06-08 | | Release date: | 2020-08-12 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody.
Science, 369, 2020
|
|
7CAK
 
 | | SARS-CoV-2 S trimer with three RBD in the open state and complexed with three H014 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of H014 Fab, ... | | Authors: | Zhe, L, Cao, L, Deng, Y, Sun, Y, Wang, N, Xie, L, Wang, Y, Rao, Z, Qin, C, Wang, X. | | Deposit date: | 2020-06-08 | | Release date: | 2020-09-23 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody.
Science, 369, 2020
|
|
9BQJ
 
 | | RO76 bound muOR-Gi1-scFv16 complex structure | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Wang, H, Majumdar, S, Kobilka, B.K. | | Deposit date: | 2024-05-10 | | Release date: | 2024-09-11 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Signaling Modulation Mediated by Ligand Water Interactions with the Sodium Site at mu OR.
Acs Cent.Sci., 10, 2024
|
|
8VLF
 
 | |
8VLD
 
 | |
8VLH
 
 | |
8CJ7
 
 | | HDAC6 selective degraded (difluoromethyl)-1,3,4-oxadiazole substrate inhibitor | | Descriptor: | 6-[(5-pyridin-2-yl-1,2$l^{4},3,4-tetrazacyclopenta-1,3-dien-2-yl)methyl]pyridine-3-carbohydrazide, Histone deacetylase 6, IODIDE ION, ... | | Authors: | Sandmark, J, Ek, M, Ripa, L. | | Deposit date: | 2023-02-12 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Selective and Bioavailable HDAC6 2-(Difluoromethyl)-1,3,4-oxadiazole Substrate Inhibitors and Modeling of Their Bioactivation Mechanism.
J.Med.Chem., 66, 2023
|
|
8W51
 
 | |
8X7T
 
 | | MCM in the Apo state. | | Descriptor: | mini-chromosome maintenance complex 3 | | Authors: | Ma, J, Yi, G, Ye, M, MacGregor-Chatwin, C, Sheng, Y, Lu, Y, Li, M, Gilbert, R.J.C, Zhang, P. | | Deposit date: | 2023-11-25 | | Release date: | 2024-01-17 | | Last modified: | 2024-12-11 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Open architecture of archaea MCM and dsDNA complexes resolved using monodispersed streptavidin affinity CryoEM.
Nat Commun, 15, 2024
|
|
8X7U
 
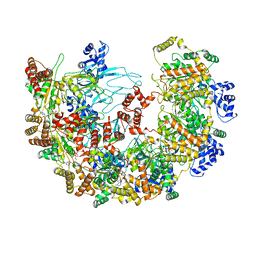 | | MCM in complex with dsDNA in presence of ATP. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ma, J, Yi, G, Ye, M, MacGregor-Chatwin, C, Sheng, Y, Lu, Y, Li, M, Gilbert, R.J.C, Zhang, P. | | Deposit date: | 2023-11-25 | | Release date: | 2024-01-17 | | Last modified: | 2025-01-01 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Open architecture of archaea MCM and dsDNA complexes resolved using monodispersed streptavidin affinity CryoEM.
Nat Commun, 15, 2024
|
|
2GHG
 
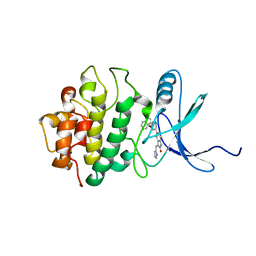 | | h-CHK1 complexed with A431994 | | Descriptor: | 5-{5-[(S)-2-AMINO-3-(1H-INDOL-3-YL)-PROPOXYL]-PYRIDIN-3-YL}-3-[1-(1H-PYRROL-2-YL)-METH-(Z)-YLIDENE]-1,3-DIHYDRO-INDOL-2-ONE, Serine/threonine-protein kinase Chk1 | | Authors: | Park, C. | | Deposit date: | 2006-03-27 | | Release date: | 2007-03-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Discovery and SAR of oxindole-pyridine-based protein kinase B/Akt inhibitors for treating cancers.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
3H00
 
 | |
3H01
 
 | |
3GWO
 
 | | Structure of the C-terminal Domain of a Putative HIV-1 gp41 Fusion Intermediate | | Descriptor: | Envelope glycoprotein gp160, O-(O-(2-AMINOPROPYL)-O'-(2-METHOXYETHYL)POLYPROPYLENE GLYCOL 500), SODIUM ION | | Authors: | Liu, J. | | Deposit date: | 2009-04-01 | | Release date: | 2009-12-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Role of a putative gp41 dimerization domain in human immunodeficiency virus type 1 membrane fusion.
J.Virol., 84, 2010
|
|
2KGL
 
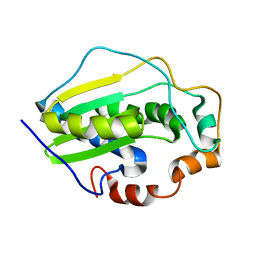 | | NMR solution structure of MESD | | Descriptor: | Mesoderm development candidate 2 | | Authors: | Chen, J, Wang, J. | | Deposit date: | 2009-03-12 | | Release date: | 2010-03-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Two Structural and Functional Domains of MESD Required for Proper Folding and Trafficking of LRP5/6.
Structure, 19, 2011
|
|
7E34
 
 | | Crystal structure of SUN1-Speedy A-CDK2 | | Descriptor: | Cyclin-dependent kinase 2, GLYCEROL, SUN domain-containing protein 1, ... | | Authors: | Chen, Y, Huang, C, Wu, J, Lei, M. | | Deposit date: | 2021-02-08 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | The SUN1-SPDYA interaction plays an essential role in meiosis prophase I.
Nat Commun, 12, 2021
|
|
8XBD
 
 | | GH18 family chitinase from cold seep metagenome | | Descriptor: | GH18 chitinase-like superfamily protein | | Authors: | Yang, J. | | Deposit date: | 2023-12-06 | | Release date: | 2023-12-27 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Exploring the role of organotrophic microbes in geochemical cycling of cold seep sediments.
Innov Geosci, 3, 2025
|
|
2L7B
 
 | | NMR Structure of full length apoE3 | | Descriptor: | Apolipoprotein E | | Authors: | Chen, J, Wang, J. | | Deposit date: | 2010-12-07 | | Release date: | 2011-08-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Topology of human apolipoprotein E3 uniquely regulates its diverse biological functions.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
5GM4
 
 | | Crystal structure of FI-CMCase from Aspergillus aculeatus F-50 in complex with cellotetrose | | Descriptor: | Endoglucanase-1, SULFATE ION, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Huang, J.W, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2016-07-12 | | Release date: | 2017-05-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure and genetic modifications of FI-CMCase from Aspergillus aculeatus F-50
Biochem. Biophys. Res. Commun., 478, 2016
|
|
5GM3
 
 | | Crystal structure of FI-CMCase from Aspergillus aculeatus F-50 | | Descriptor: | CACODYLATE ION, Endoglucanase-1, ZINC ION | | Authors: | Huang, J.W, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2016-07-12 | | Release date: | 2017-05-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Crystal structure and genetic modifications of FI-CMCase from Aspergillus aculeatus F-50
Biochem. Biophys. Res. Commun., 478, 2016
|
|
5GM9
 
 | | Crystal structure of a glycoside hydrolase in complex with cellobiose | | Descriptor: | Glycoside hydrolase family 45 protein, beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Gao, J, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2016-07-13 | | Release date: | 2017-04-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Characterization and crystal structure of a thermostable glycoside hydrolase family 45 1,4-beta-endoglucanase from Thielavia terrestris
Enzyme Microb. Technol., 99, 2017
|
|
5H31
 
 | |
2MRI
 
 | |
2MR3
 
 | | A subunit of 26S proteasome lid complex | | Descriptor: | 26S proteasome regulatory subunit RPN9 | | Authors: | Wu, Y, Hu, Y, Jin, C. | | Deposit date: | 2014-06-30 | | Release date: | 2015-02-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of yeast Rpn9: insights into proteasome lid assembly.
J. Biol. Chem., 290, 2015
|
|
2MQW
 
 | |
