5ZYA
 
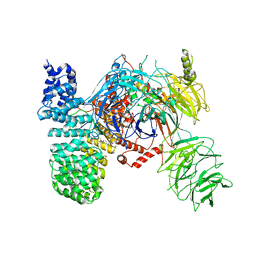 | | SF3b spliceosomal complex bound to E7107 | | Descriptor: | PHD finger-like domain-containing protein 5A, POTASSIUM ION, Splicing factor 3B subunit 1, ... | | Authors: | Finci, L.I, Larsen, N.A. | | Deposit date: | 2018-05-23 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | The cryo-EM structure of the SF3b spliceosome complex bound to a splicing modulator reveals a pre-mRNA substrate competitive mechanism of action
Genes Dev., 32, 2018
|
|
8TGW
 
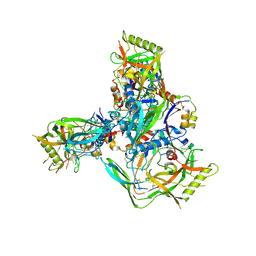 | | Cryo-EM structure of 1059 SOSIP trimer purified via Galanthus nivalis lectin chromatography | | Descriptor: | 1059 SOSIP Surface protein gp120, 1059 SOSIP Transmembrane protein gp41, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Parsons, R.J, Pothula, K, Acharya, P. | | Deposit date: | 2023-07-13 | | Release date: | 2024-08-28 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Conformational flexibility of HIV-1 envelope glycoproteins modulates transmitted/founder sensitivity to broadly neutralizing antibodies.
Nat Commun, 15, 2024
|
|
8TGU
 
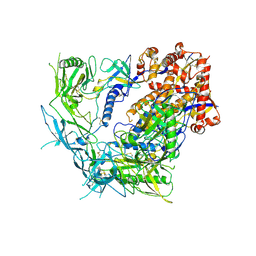 | | Cryo-EM structure of BG505 SOSIP trimer purified via Galanthus nivalis lectin chromatography | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 Envelope glycoprotein BG505 SOSIP, ... | | Authors: | Parsons, R.J, Pothula, K, Acharya, P. | | Deposit date: | 2023-07-13 | | Release date: | 2024-08-28 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Conformational flexibility of HIV-1 envelope glycoproteins modulates transmitted/founder sensitivity to broadly neutralizing antibodies.
Nat Commun, 15, 2024
|
|
8G8C
 
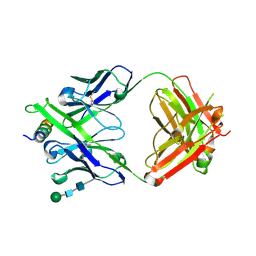 | | Crystal structure of DH1322.1 Fab in complex with HIV proximal MPER peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DH1322.1 heavy chain, DH1322.1 light chain, ... | | Authors: | Niyongabo, A, Janus, B.M, Ofek, G. | | Deposit date: | 2023-02-17 | | Release date: | 2024-05-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Vaccine induction of heterologous HIV-1-neutralizing antibody B cell lineages in humans.
Cell, 187, 2024
|
|
8U89
 
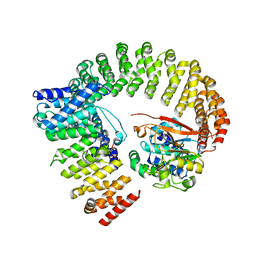 | | The structure of the PP2A-B56Delta holoenzyme mutant - E197K | | Descriptor: | MANGANESE (II) ION, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit delta isoform, Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform, ... | | Authors: | Wu, C.G, Xing, Y. | | Deposit date: | 2023-09-16 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | B56 delta long-disordered arms form a dynamic PP2A regulation interface coupled with global allostery and Jordan's syndrome mutations.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8H7V
 
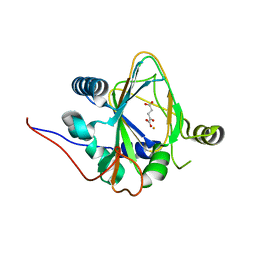 | | Trans-3/4-proline-hydroxylase H11 with AKG | | Descriptor: | 2-OXOGLUTARIC ACID, FE (III) ION, Phytanoyl-CoA dioxygenase | | Authors: | Gong, W.M, Hu, X.Y. | | Deposit date: | 2022-10-21 | | Release date: | 2023-04-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structures of L-proline trans-hydroxylase reveal the catalytic specificity and provide deeper insight into AKG-dependent hydroxylation.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8H85
 
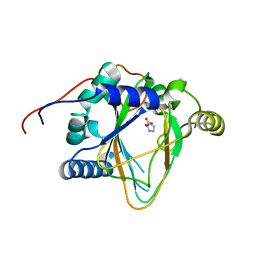 | | Trans-3/4-proline-hydroxylase H11 with 3-hydroxyl-proline | | Descriptor: | 3-HYDROXYPROLINE, Phytanoyl-CoA dioxygenase | | Authors: | Gong, W.M, Hu, X.Y. | | Deposit date: | 2022-10-21 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structures of L-proline trans-hydroxylase reveal the catalytic specificity and provide deeper insight into AKG-dependent hydroxylation.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8H81
 
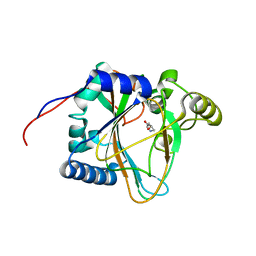 | | Trans-3/4-proline-hydroxylase H11 with 4-Hydroxyl-proline | | Descriptor: | 4-HYDROXYPROLINE, Phytanoyl-CoA dioxygenase | | Authors: | Gong, W.M, Hu, X.Y. | | Deposit date: | 2022-10-21 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structures of L-proline trans-hydroxylase reveal the catalytic specificity and provide deeper insight into AKG-dependent hydroxylation.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8H7T
 
 | | Trans-3/4-proline-hydroxylase H11 apo structure | | Descriptor: | CHLORIDE ION, Phytanoyl-CoA dioxygenase | | Authors: | Gong, W.M, Hu, X.Y. | | Deposit date: | 2022-10-21 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structures of L-proline trans-hydroxylase reveal the catalytic specificity and provide deeper insight into AKG-dependent hydroxylation.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8H7Y
 
 | | Trans-3/4-proline-hydroxylase H11 with AKG and L-proline | | Descriptor: | 2-OXOGLUTARIC ACID, FE (III) ION, PROLINE, ... | | Authors: | Gong, W.M, Hu, X.Y. | | Deposit date: | 2022-10-21 | | Release date: | 2023-04-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structures of L-proline trans-hydroxylase reveal the catalytic specificity and provide deeper insight into AKG-dependent hydroxylation.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
6JFP
 
 | | Crystal structure of the beta-glucosidase Bgl15 | | Descriptor: | beta-D-glucopyranose, beta-glucosidase 15 | | Authors: | Xie, W, Chen, R. | | Deposit date: | 2019-02-11 | | Release date: | 2020-02-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Engineering of beta-Glucosidase Bgl15 with Simultaneously Enhanced Glucose Tolerance and Thermostability To Improve Its Performance in High-Solid Cellulose Hydrolysis.
J.Agric.Food Chem., 68, 2020
|
|
8IFP
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound 1 | | Descriptor: | (1R,2S,5S)-3-[(2S)-2-(tert-butylcarbamoylamino)-3,3-dimethyl-butanoyl]-6,6-dimethyl-N-[(2S)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Nie, T.Q, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-02-19 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure-based development and preclinical evaluation of the SARS-CoV-2 3C-like protease inhibitor simnotrelvir.
Nat Commun, 14, 2023
|
|
8IFT
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound 10 | | Descriptor: | (8S)-N-[(1S)-1-cyano-2-[(3S)-2-oxidanylidenepyrrolidin-3-yl]ethyl]-7-[(2S)-2-[(1-fluoranylcyclopropyl)carbonylamino]-3,3-dimethyl-butanoyl]-1,4-dithia-7-azaspiro[4.4]nonane-8-carboxamide, 3C-like proteinase nsp5 | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Nie, T.Q, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-02-19 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based development and preclinical evaluation of the SARS-CoV-2 3C-like protease inhibitor simnotrelvir.
Nat Commun, 14, 2023
|
|
8IFQ
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound 2 | | Descriptor: | (1R,2S,5S)-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-N-{(1Z,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Nie, T.Q, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-02-19 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure-based development and preclinical evaluation of the SARS-CoV-2 3C-like protease inhibitor simnotrelvir.
Nat Commun, 14, 2023
|
|
8IFS
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound 7 | | Descriptor: | (8~{S})-7-[(2~{S})-2-(~{tert}-butylcarbamoylamino)-3,3-dimethyl-butanoyl]-~{N}-[(1~{S})-1-cyano-2-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]ethyl]-1,4-dithia-7-azaspiro[4.4]nonane-8-carboxamide, 3C-like proteinase nsp5 | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Nie, T.Q, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-02-19 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structure-based development and preclinical evaluation of the SARS-CoV-2 3C-like protease inhibitor simnotrelvir.
Nat Commun, 14, 2023
|
|
8IFR
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound 3 | | Descriptor: | (1R,2S,5S)-3-[(2S)-2-(tert-butylcarbamoylamino)-3,3-dimethyl-butanoyl]-6,6-dimethyl-N-[(2S)-5-oxidanylidene-1-[(3S)-2-oxidanylidenepyrrolidin-3-yl]hex-3-en-2-yl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Nie, T.Q, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-02-19 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structure-based development and preclinical evaluation of the SARS-CoV-2 3C-like protease inhibitor simnotrelvir.
Nat Commun, 14, 2023
|
|
8IGX
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound 9 (simnotrelvir, SIM0417, SSD8432) | | Descriptor: | (8~{S})-~{N}-[(1~{S})-1-cyano-2-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]ethyl]-7-[(2~{S})-3,3-dimethyl-2-[2,2,2-tris(fluoranyl)ethanoylamino]butanoyl]-1,4-dithia-7-azaspiro[4.4]nonane-8-carboxamide, 3C-like proteinase nsp5 | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Nie, T.Q, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-02-21 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based development and preclinical evaluation of the SARS-CoV-2 3C-like protease inhibitor simnotrelvir.
Nat Commun, 14, 2023
|
|
8IGY
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Nie, T.Q, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-02-21 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure-based development and preclinical evaluation of the SARS-CoV-2 3C-like protease inhibitor simnotrelvir.
Nat Commun, 14, 2023
|
|
7KN4
 
 | |
8VLT
 
 | |
6J69
 
 | | Structure of KIBRA and Dendrin Complex | | Descriptor: | Peptide from Dendrin, Protein KIBRA | | Authors: | Lin, Z, Yang, Z, Ji, Z, Zhang, M. | | Deposit date: | 2019-01-14 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.753 Å) | | Cite: | Kibra Modulates Learning and Memory via Binding to Dendrin.
Cell Rep, 26, 2019
|
|
7FAJ
 
 | | CARM1 bound with compound 43 | | Descriptor: | Histone-arginine methyltransferase CARM1, N'-[[3-[4-(3,5-dimethyl-1,2-oxazol-4-yl)-5-methyl-6-phenylazanyl-pyrimidin-2-yl]phenyl]methyl]-N-methyl-ethane-1,2-diamine | | Authors: | Cao, D.Y, Li, J, Xiong, B. | | Deposit date: | 2021-07-06 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2450726 Å) | | Cite: | Structure-Based Discovery of Potent CARM1 Inhibitors for Solid Tumor and Cancer Immunology Therapy.
J.Med.Chem., 64, 2021
|
|
7FAI
 
 | | CARM1 bound with compound 9 | | Descriptor: | Histone-arginine methyltransferase CARM1, N'-[[3-[4-(3,5-dimethyl-1,2-oxazol-4-yl)-5-methyl-6-(oxan-4-ylamino)pyrimidin-2-yl]phenyl]methyl]-N-methyl-ethane-1,2-diamine | | Authors: | Cao, D.Y, Li, J, Xiong, B. | | Deposit date: | 2021-07-06 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09749269 Å) | | Cite: | Structure-Based Discovery of Potent CARM1 Inhibitors for Solid Tumor and Cancer Immunology Therapy.
J.Med.Chem., 64, 2021
|
|
6XAF
 
 | | 1.9A crystal structure of the GTPase domain of Parkinson's disease-associated protein LRRK2 carrying R1398H | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Leucine-rich repeat serine/threonine-protein kinase 2, MAGNESIUM ION | | Authors: | Hoang, Q.Q, Liao, J, Huang, X, Park, Y, Wu, C.X. | | Deposit date: | 2020-06-04 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.968 Å) | | Cite: | Structural basis for conformational plasticity in the GTPase domain of the Parkinson's disease-associated protein LRRK2
To be Published
|
|
7UB0
 
 | |
