4NNI
 
 | | Structural basis for targeting the ribosomal protein S1 of Mycobacterium tuberculosis by pyrazinamide | | Descriptor: | 30S ribosomal protein S1, PYRAZINE-2-CARBOXYLIC ACID | | Authors: | Yang, J, Liu, Y, Cai, Q, Lin, D. | | Deposit date: | 2013-11-18 | | Release date: | 2014-12-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structural basis for targeting the ribosomal protein S1 of Mycobacterium tuberculosis by pyrazinamide.
Mol.Microbiol., 95, 2015
|
|
6JHJ
 
 | | Structure of Marine bacterial laminarinase mutant-E135A | | Descriptor: | CALCIUM ION, LamCAT | | Authors: | Yang, J, Xu, Y, Miyakawa, T, Tanokura, M, Long, L. | | Deposit date: | 2019-02-18 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Molecular Basis for Substrate Recognition and Catalysis by a Marine Bacterial Laminarinase.
Appl.Environ.Microbiol., 86, 2020
|
|
4NNK
 
 | |
1ASH
 
 | | THE STRUCTURE OF ASCARIS HEMOGLOBIN DOMAIN I AT 2.2 ANGSTROMS RESOLUTION: MOLECULAR FEATURES OF OXYGEN AVIDITY | | Descriptor: | HEMOGLOBIN (OXY), OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yang, J, Mathews, F.S, Kloek, A.P, Goldberg, D.E. | | Deposit date: | 1995-01-06 | | Release date: | 1995-02-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The structure of Ascaris hemoglobin domain I at 2.2 A resolution: molecular features of oxygen avidity.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
2ASI
 
 | | ASPARTIC PROTEINASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ASPARTIC PROTEINASE, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yang, J, Jia, Z, Vandonselaar, M, Kepliakov, P.S.A, Quail, J.W. | | Deposit date: | 1995-12-09 | | Release date: | 1996-08-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of the aspartic proteinase from Rhizomucor miehei at 2.15 A resolution.
J.Mol.Biol., 268, 1997
|
|
6LPX
 
 | | Crystal structure of human D-2-hydroxyglutarate dehydrogenase in complex with 2-oxoglutarate (2-OG) | | Descriptor: | 2-OXOGLUTARIC ACID, D-2-hydroxyglutarate dehydrogenase, mitochondrial, ... | | Authors: | Yang, J, Zhu, H, Ding, J. | | Deposit date: | 2020-01-12 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure, substrate specificity, and catalytic mechanism of human D-2-HGDH and insights into pathogenicity of disease-associated mutations.
Cell Discov, 7, 2021
|
|
6LPQ
 
 | | Crystal structure of human D-2-hydroxyglutarate dehydrogenase in complex with D-malate (D-MAL) | | Descriptor: | D-2-hydroxyglutarate dehydrogenase, mitochondrial, D-MALATE, ... | | Authors: | Yang, J, Zhu, H, Ding, J. | | Deposit date: | 2020-01-12 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure, substrate specificity, and catalytic mechanism of human D-2-HGDH and insights into pathogenicity of disease-associated mutations.
Cell Discov, 7, 2021
|
|
6LPN
 
 | | Crystal structure of human D-2-hydroxyglutarate dehydrogenase in apo form | | Descriptor: | D-2-hydroxyglutarate dehydrogenase, mitochondrial, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Yang, J, Zhu, H, Ding, J. | | Deposit date: | 2020-01-12 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Structure, substrate specificity, and catalytic mechanism of human D-2-HGDH and insights into pathogenicity of disease-associated mutations.
Cell Discov, 7, 2021
|
|
6LPT
 
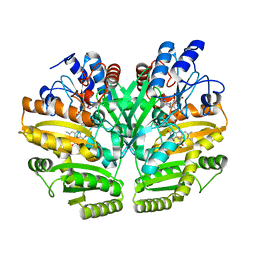 | | Crystal structure of human D-2-hydroxyglutarate dehydrogenase in complex with D-lactate (D-LAC) | | Descriptor: | D-2-hydroxyglutarate dehydrogenase, mitochondrial, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Yang, J, Zhu, H, Ding, J. | | Deposit date: | 2020-01-12 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.621 Å) | | Cite: | Structure, substrate specificity, and catalytic mechanism of human D-2-HGDH and insights into pathogenicity of disease-associated mutations.
Cell Discov, 7, 2021
|
|
6LPP
 
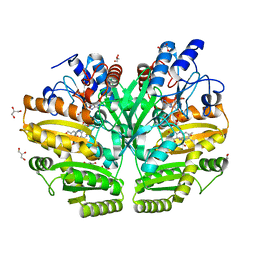 | | Crystal structure of human D-2-hydroxyglutarate dehydrogenase in complex with D-2-hydroxyglutarate (D-2-HG) | | Descriptor: | (2R)-2-hydroxypentanedioic acid, D-2-hydroxyglutarate dehydrogenase, mitochondrial, ... | | Authors: | Yang, J, Zhu, H, Ding, J. | | Deposit date: | 2020-01-12 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure, substrate specificity, and catalytic mechanism of human D-2-HGDH and insights into pathogenicity of disease-associated mutations.
Cell Discov, 7, 2021
|
|
6LPU
 
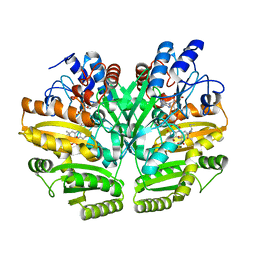 | | Crystal structure of human D-2-hydroxyglutarate dehydrogenase in complex with L-2-hydroxyglutarate (L-2-HG) | | Descriptor: | (2S)-2-HYDROXYPENTANEDIOIC ACID, D-2-hydroxyglutarate dehydrogenase, mitochondrial, ... | | Authors: | Yang, J, Zhu, H, Ding, J. | | Deposit date: | 2020-01-12 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.923 Å) | | Cite: | Structure, substrate specificity, and catalytic mechanism of human D-2-HGDH and insights into pathogenicity of disease-associated mutations.
Cell Discov, 7, 2021
|
|
8J0P
 
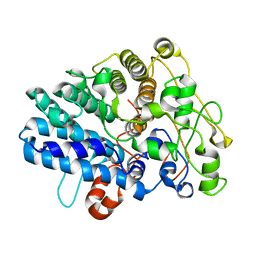 | | Chitin binding SusD-like protein AqSusD from a marine Bacteroidetes | | Descriptor: | Chitin binding SusD-like protein | | Authors: | Yang, J. | | Deposit date: | 2023-04-11 | | Release date: | 2023-11-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights of a SusD-like protein in marine Bacteroidetes bacteria reveal the molecular basis for chitin recognition and acquisition.
Febs J., 291, 2024
|
|
8JXZ
 
 | | Chitin binding SusD-like protein AqSusD in complex with (GlcNAc)3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SusD-like protein AqSusD | | Authors: | Yang, J. | | Deposit date: | 2023-07-01 | | Release date: | 2023-11-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights of a SusD-like protein in marine Bacteroidetes bacteria reveal the molecular basis for chitin recognition and acquisition.
Febs J., 291, 2024
|
|
2B3O
 
 | | Crystal structure of human tyrosine phosphatase SHP-1 | | Descriptor: | Tyrosine-protein phosphatase, non-receptor type 6 | | Authors: | Yang, J, Liu, L, He, D, Song, X, Liang, X, Zhao, Z.J, Zhou, G.W. | | Deposit date: | 2005-09-20 | | Release date: | 2005-10-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of human protein-tyrosine phosphatase SHP-1.
J.Biol.Chem., 278, 2003
|
|
7W1S
 
 | | Crystal structure of SARS-CoV-2 spike receptor-binding domain in complex with neutralizing nanobody Nb-007 | | Descriptor: | Nanobody Nb-007, Spike protein S1 | | Authors: | Yang, J, Lin, S, Sun, H.L, Lu, G.W. | | Deposit date: | 2021-11-20 | | Release date: | 2022-06-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | A Potent Neutralizing Nanobody Targeting the Spike Receptor-Binding Domain of SARS-CoV-2 and the Structural Basis of Its Intimate Binding.
Front Immunol, 13, 2022
|
|
7VQM
 
 | | GH2 beta-galacturonate AqGalA in complex with galacturonide | | Descriptor: | CHLORIDE ION, GH2 beta-galacturonate AqGalA, beta-D-galactopyranuronic acid | | Authors: | Yang, J. | | Deposit date: | 2021-10-20 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Biochemical Basis of a Marine Bacterial Glycoside Hydrolase Family 2 beta-Glycosidase with Broad Substrate Specificity
Appl.Environ.Microbiol., 88, 2022
|
|
6M6P
 
 | | Structure of Marine bacterial laminarinase mutant E135A in complex with 1,3-beta-cellotriosyl-glucose | | Descriptor: | CALCIUM ION, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-3)-alpha-D-glucopyranose, laminarinase | | Authors: | Yang, J, Xu, Y, Tanokura, M, Long, L, Miyakawa, T. | | Deposit date: | 2020-03-16 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Molecular Basis for Substrate Recognition and Catalysis by a Marine Bacterial Laminarinase.
Appl.Environ.Microbiol., 86, 2020
|
|
6NRY
 
 | |
6NS7
 
 | |
2LR7
 
 | | Cathelicidin-PY | | Descriptor: | Cathelicidin-PY | | Authors: | Yang, J. | | Deposit date: | 2012-03-27 | | Release date: | 2013-03-27 | | Method: | SOLUTION NMR | | Cite: | Structure of Cathelicidin-PY
To be Published
|
|
7VW2
 
 | |
7VW1
 
 | |
7VW0
 
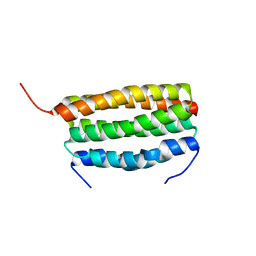 | | Structure of a dimeric periplasmic protein | | Descriptor: | DUF305 domain-containing protein | | Authors: | Yang, J, Liu, L. | | Deposit date: | 2021-11-09 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.447 Å) | | Cite: | Structural basis of copper binding by a dimeric periplasmic protein forming a six-helical bundle.
J.Inorg.Biochem., 229, 2022
|
|
8K2P
 
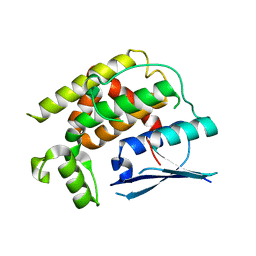 | | Crystal structure of CtGST-F76A | | Descriptor: | Glutathione S-transferase | | Authors: | Yang, J, Xiao, J.Y, Lei, X.G. | | Deposit date: | 2023-07-13 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Enzymatic Degradation of Deoxynivalenol with the Engineered Detoxification Enzyme Fhb7.
Jacs Au, 4, 2024
|
|
8K2O
 
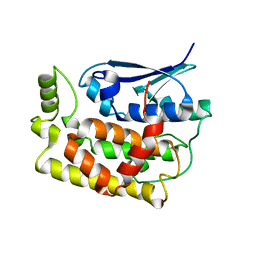 | | Crystal structure of Fhb7-M10 | | Descriptor: | Fhb7-M10 | | Authors: | Yang, J, Lei, X.G, Xiao, J.Y. | | Deposit date: | 2023-07-13 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Enzymatic Degradation of Deoxynivalenol with the Engineered Detoxification Enzyme Fhb7.
Jacs Au, 4, 2024
|
|
