6L9L
 
 | | 1D4 TCR recognition of H2-Ld a1a2 A5 Peptide Complexes | | Descriptor: | H2-Ld a1a2, SER-PRO-SER-TYR-ALA-TYR-HIS-GLN-PHE, T Cell Receptor | | Authors: | Wei, P.C, Yin, L. | | Deposit date: | 2019-11-10 | | Release date: | 2020-11-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Structures suggest an approach for converting weak self-peptide tumor antigens into superagonists for CD8 T cells in cancer.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6L9M
 
 | | H2-Ld complexed with AH1 peptide | | Descriptor: | H2-Ld, SER-PRO-SER-TYR-VAL-TYR-HIS-GLN-PHE, b2m | | Authors: | Wei, P.C, Yin, L. | | Deposit date: | 2019-11-10 | | Release date: | 2020-11-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures suggest an approach for converting weak self-peptide tumor antigens into superagonists for CD8 T cells in cancer.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5XS3
 
 | | Crystal structure of HLA Class I antigen | | Descriptor: | Heavy Chain, Light Chain, P | | Authors: | Wei, P.C, Yang, Y, Liu, Z.X, Luo, Z.Q, Tu, W.Y, Han, J.Y, Deng, Y.H, Yin, L. | | Deposit date: | 2017-06-12 | | Release date: | 2017-09-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Characterization of Autoantigen Presentation by HLA-C*06:02 in Psoriasis
J. Invest. Dermatol., 137, 2017
|
|
6JQ3
 
 | | Crystal Structure of H2-Kb in complex with a DPAGT1 mutant peptide | | Descriptor: | Beta-2-microglobulin, DPAGT1 mutant antigen SIIVFNLL, H-2 class I histocompatibility antigen, ... | | Authors: | Bai, P, Zhou, Q, Wei, P, Yin, L. | | Deposit date: | 2019-03-28 | | Release date: | 2020-05-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Immune-based mutation classification enables neoantigen prioritization and immune feature discovery in cancer immunotherapy.
Oncoimmunology, 10, 2021
|
|
6JTN
 
 | | Crystal structure of HLA-C08 in complex with a tumor mut10m peptide | | Descriptor: | 10-mer peptide, Beta-2-microglobulin, HLA class I antigen, ... | | Authors: | Bai, P, Zhou, Q, Wei, P, Yin, L. | | Deposit date: | 2019-04-11 | | Release date: | 2020-04-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Immune-based mutation classification enables neoantigen prioritization and immune feature discovery in cancer immunotherapy.
Oncoimmunology, 10, 2021
|
|
6JTP
 
 | | Crystal structure of HLA-C08 in complex with a tumor mut9m peptide | | Descriptor: | 9-mer peptide, Beta-2-microglobulin, HLA class I antigen, ... | | Authors: | Bai, P, Zhou, Q, Wei, P, Lei, Y. | | Deposit date: | 2019-04-11 | | Release date: | 2020-04-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Immune-based mutation classification enables neoantigen prioritization and immune feature discovery in cancer immunotherapy.
Oncoimmunology, 10, 2021
|
|
8U77
 
 | | Crystal structure of Taf14 in complex with Yng1 | | Descriptor: | Protein YNG1, Transcription initiation factor TFIID subunit 14 | | Authors: | Nguyen, M.C, Wei, P.C, Zhang, G.Y, Kutateladze, T.G. | | Deposit date: | 2023-09-14 | | Release date: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Molecular insight into interactions between the Taf14, Yng1 and Sas3 subunits of the NuA3 complex.
Nat Commun, 15, 2024
|
|
7PIZ
 
 | |
7PJC
 
 | |
7PPR
 
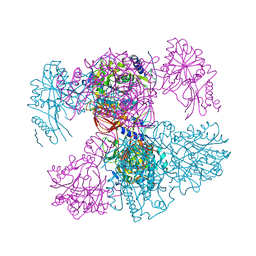 | |
7R7R
 
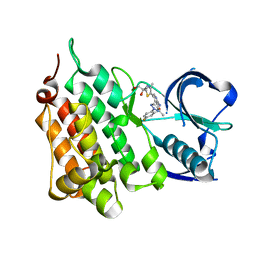 | | Structure of Human Anaplastic Lymphoma Kinase Domain in complex with ((2~{R})-2-[5-[6-amino-5-[(1~{R})-1-[5-fluoro-2-(triazol-2-yl)phenyl]ethoxy]-3-pyridyl]-4-methyl-thiazol-2-yl]propane-1,2-diol) | | Descriptor: | (2R)-2-[5-(6-amino-5-{(1R)-1-[2-(1,3-dihydro-2H-1,2,3-triazol-2-yl)-5-fluorophenyl]ethoxy}pyridin-3-yl)-4-methyl-1,3-thiazol-2-yl]propane-1,2-diol, ALK tyrosine kinase receptor | | Authors: | McTigue, M. | | Deposit date: | 2021-06-25 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.935 Å) | | Cite: | Analysis of lorlatinib analogs reveals a roadmap for targeting diverse compound resistance mutations in ALK-positive lung cancer.
Nat Cancer, 3, 2022
|
|
7R7K
 
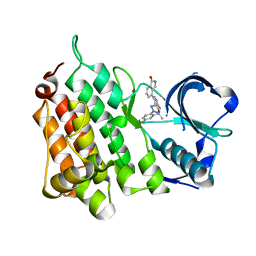 | |
8KHD
 
 | | The interface structure of Omicron RBD binding to 5817 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 5817, Light chain of 5817, ... | | Authors: | Cao, L, Wang, X. | | Deposit date: | 2023-08-21 | | Release date: | 2024-04-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Identification of a broad sarbecovirus neutralizing antibody targeting a conserved epitope on the receptor-binding domain.
Cell Rep, 43, 2024
|
|
8KHC
 
 | | SARS-CoV-2 Omicron spike in complex with 5817 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 5817 Fab, ... | | Authors: | Cao, L, Wang, X. | | Deposit date: | 2023-08-21 | | Release date: | 2024-04-17 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Identification of a broad sarbecovirus neutralizing antibody targeting a conserved epitope on the receptor-binding domain.
Cell Rep, 43, 2024
|
|
8H0R
 
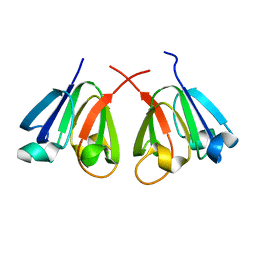 | |
8IVL
 
 | | FABP7 complexed with Cholesterol | | Descriptor: | CHOLESTEROL, Fatty acid-binding protein, brain | | Authors: | Wei, P.C, Zhao, K, Yin, L. | | Deposit date: | 2023-03-28 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Fatty acid-binding proteins 3, 7, and 8 bind cholesterol and facilitate its egress from lysosomes.
J.Cell Biol., 223, 2024
|
|
8IVF
 
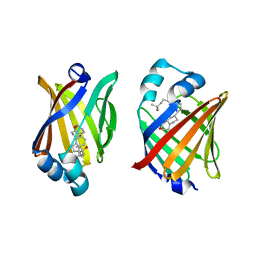 | | FABP7 complexed with 25-HC | | Descriptor: | 25-HYDROXYCHOLESTEROL, Fatty acid-binding protein, brain | | Authors: | Wei, P.C, Zhao, K, Yin, L. | | Deposit date: | 2023-03-27 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Fatty acid-binding proteins 3, 7, and 8 bind cholesterol and facilitate its egress from lysosomes.
J.Cell Biol., 223, 2024
|
|
6F1J
 
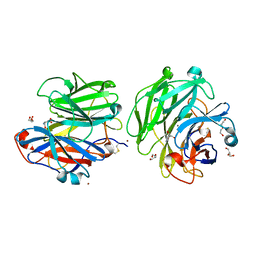 | | Structure of a Talaromyces pinophilus GH62 Arabinofuranosidase in complex with AraDNJ at 1.25A resolution | | Descriptor: | 1,4-DIDEOXY-1,4-IMINO-L-ARABINITOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Moroz, O.V, Sobala, L, Blagova, E, Coyle, T, Morkeberg Krogh, K.B.R, Wei, P, Stubbs, K, Wilson, K.S, Davies, G.J. | | Deposit date: | 2017-11-22 | | Release date: | 2018-08-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structure of a Talaromyces pinophilus GH62 arabinofuranosidase in complex with AraDNJ at 1.25 angstrom resolution.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
5Z0S
 
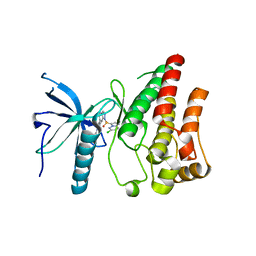 | | Crystal structure of FGFR1 kinase domain in complex with a novel inhibitor | | Descriptor: | 1-[(6-chloroimidazo[1,2-b]pyridazin-3-yl)sulfonyl]-6-(1-methyl-1H-pyrazol-4-yl)-1H-pyrazolo[4,3-b]pyridine, Fibroblast growth factor receptor 1 | | Authors: | Liu, Q, Xu, Y. | | Deposit date: | 2017-12-20 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure-Based Discovery of a Series of 5H-Pyrrolo[2,3-b]pyrazine FGFR Kinase Inhibitors
Molecules, 23, 2018
|
|
8YZS
 
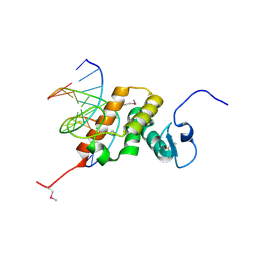 | | Structure of the NACC1 BEN domain in complex with its target DNA | | Descriptor: | CATG-containing DNA, Nucleus accumbens-associated protein 1 | | Authors: | Ren, J, Wang, Z. | | Deposit date: | 2024-04-08 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural basis of DNA recognition by BEN domain proteins reveals a role for oligomerization in unmethylated DNA selection by BANP.
Nucleic Acids Res., 52, 2024
|
|
8YZT
 
 | |
9BLG
 
 | | Crystal structure of non-receptor protein tyrosine phosphatase SHP2 in complex with PF-07284892 | | Descriptor: | (1S)-1'-{6-[(2-amino-3-chloropyridin-4-yl)sulfanyl]-1,2,4-triazin-3-yl}-1,3-dihydrospiro[indene-2,4'-piperidin]-1-amine, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Bester, S.M, Wu, W.-I, Mou, T.-C. | | Deposit date: | 2024-04-30 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | SHP2 Inhibition Sensitizes Diverse Oncogene-Addicted Solid Tumors to Re-treatment with Targeted Therapy.
Cancer Discov, 13, 2023
|
|
7DUU
 
 | | Crystal structure of HLA molecule with an KIR receptor | | Descriptor: | Beta-2-microglobulin, Killer cell immunoglobulin-like receptor 2DS2, LEU-ASN-PRO-SER-VAL-ALA-ALA-THR-LEU, ... | | Authors: | Yang, Y, Yin, L. | | Deposit date: | 2021-01-11 | | Release date: | 2022-02-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Activating receptor KIR2DS2 bound to HLA-C1 reveals the novel recognition features of activating receptor.
Immunology, 165, 2022
|
|
7W9K
 
 | | Cryo-EM structure of human Nav1.7-beta1-beta2 complex at 2.2 angstrom resolution | | Descriptor: | (2S,3R,4E)-2-(acetylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Yan, N, Huang, G, Liu, D, Wei, P, Shen, H. | | Deposit date: | 2021-12-09 | | Release date: | 2022-06-01 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | High-resolution structures of human Na v 1.7 reveal gating modulation through alpha-pi helical transition of S6 IV.
Cell Rep, 39, 2022
|
|
7W9M
 
 | | Cryo-EM structure of human Nav1.7(E406K) in complex with auxiliary beta subunits, ProTx-II and tetrodotoxin (S6IV pi helix conformer) | | Descriptor: | (1R,5R,6R,7R,9S,11S,12S,13S,14S)-3-amino-14-(hydroxymethyl)-8,10-dioxa-2,4-diazatetracyclo[7.3.1.1~7,11~.0~1,6~]tetradec-3-ene-5,9,12,13,14-pentol (non-preferred name), 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, N, Huang, G, Liu, D, Wei, P, Shen, H. | | Deposit date: | 2021-12-10 | | Release date: | 2022-05-25 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | High-resolution structures of human Na v 1.7 reveal gating modulation through alpha-pi helical transition of S6 IV.
Cell Rep, 39, 2022
|
|
