2LZF
 
 | |
5CYS
 
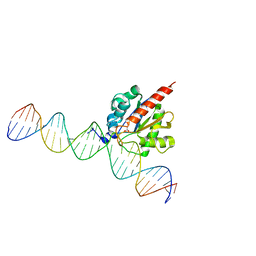 | |
2Q91
 
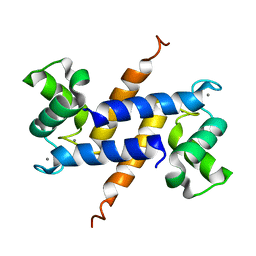 | | Structure of the Ca2+-Bound Activated Form of the S100A4 Metastasis Factor | | Descriptor: | CALCIUM ION, S100A4 Metastasis Factor | | Authors: | Malashkevich, V.N, Knight, D, Ramagopal, U.A, Almo, S.C, Bresnick, A.R. | | Deposit date: | 2007-06-12 | | Release date: | 2008-02-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structure of Ca(2+)-Bound S100A4 and Its Interaction with Peptides Derived from Nonmuscle Myosin-IIA.
Biochemistry, 47, 2008
|
|
6UWI
 
 | | Crystal structure of the Clostridium difficile translocase CDTb | | Descriptor: | ADP-ribosyltransferase binding component, CALCIUM ION | | Authors: | Pozharski, E. | | Deposit date: | 2019-11-05 | | Release date: | 2020-01-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structure of the cell-binding component of theClostridium difficilebinary toxin reveals a di-heptamer macromolecular assembly.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6UWR
 
 | |
6UWO
 
 | |
6UWT
 
 | |
2K7O
 
 | | Ca2+-S100B, refined with RDCs | | Descriptor: | CALCIUM ION, Protein S100-B | | Authors: | Wright, N.T, Inman, K.G, Levine, J.A, Weber, D.J. | | Deposit date: | 2008-08-17 | | Release date: | 2008-11-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Refinement of the solution structure and dynamic properties of Ca(2+)-bound rat S100B.
J.Biomol.Nmr, 42, 2008
|
|
3KO0
 
 | | Structure of the tfp-ca2+-bound activated form of the s100a4 Metastasis factor | | Descriptor: | 10-[3-(4-METHYL-PIPERAZIN-1-YL)-PROPYL]-2-TRIFLUOROMETHYL-10H-PHENOTHIAZINE, CALCIUM ION, Protein S100-A4 | | Authors: | Malashkevich, V.N, Dulyaninova, N.G, Knight, D, Almo, S.C, Bresnick, A.R. | | Deposit date: | 2009-11-12 | | Release date: | 2010-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Phenothiazines inhibit S100A4 function by inducing protein oligomerization.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3LLE
 
 | | X-ray structure of bovine SC0322,Ca(2+)-S100B | | Descriptor: | 13-methyl-13,14-dihydro[1,3]benzodioxolo[5,6-c][1,3]dioxolo[4,5-i]phenanthridine, CALCIUM ION, Protein S100-B | | Authors: | Charpentier, T.H, Weber, D.J, Wilder, P.W. | | Deposit date: | 2010-01-28 | | Release date: | 2010-12-29 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | In vitro screening and structural characterization of inhibitors of the S100B-p53 interaction.
Int.J.High Throughput Screen, 2010, 2010
|
|
3LK1
 
 | | X-ray structure of bovine SC0322,Ca(2+)-S100B | | Descriptor: | 2-sulfanylbenzoic acid, CALCIUM ION, ETHYL MERCURY ION, ... | | Authors: | Charpentier, T.H, Weber, D.J, Wilder, P.W. | | Deposit date: | 2010-01-26 | | Release date: | 2010-12-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | In vitro screening and structural characterization of inhibitors of the S100B-p53 interaction.
Int J High Throughput Screen, 2010, 2010
|
|
3LK0
 
 | | X-ray structure of bovine SC0067,Ca(2+)-S100B | | Descriptor: | 3-(2-chloro-10H-phenothiazin-10-yl)-N,N-dimethylpropan-1-amine, CALCIUM ION, Protein S100-B | | Authors: | Charpentier, T.H, Weber, D.J, Wilder, P.W. | | Deposit date: | 2010-01-26 | | Release date: | 2010-12-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | In vitro screening and structural characterization of inhibitors of the S100B-p53 interaction.
Int J High Throughput Screen, 2010, 2010
|
|
3RM1
 
 | |
5JXY
 
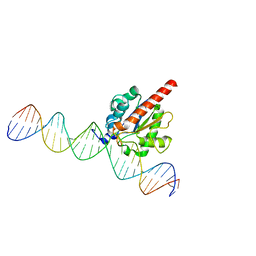 | | Enzyme-substrate complex of TDG catalytic domain bound to a G/U analog | | Descriptor: | DNA (28-MER), G/T mismatch-specific thymine DNA glycosylase | | Authors: | Pidugu, L.S, Pozharski, E, Malik, S.S, Drohat, A.C. | | Deposit date: | 2016-05-13 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structural basis of damage recognition by thymine DNA glycosylase: Key roles for N-terminal residues.
Nucleic Acids Res., 44, 2016
|
|
4Z47
 
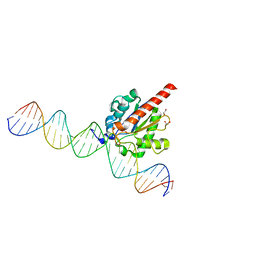 | | Structure of the enzyme-product complex resulting from TDG action on a GU mismatch in the presence of excess base | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, DNA, ... | | Authors: | Pozharski, E, Malik, S.S, Drohat, A.C. | | Deposit date: | 2015-04-01 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Thymine DNA glycosylase exhibits negligible affinity for nucleobases that it removes from DNA.
Nucleic Acids Res., 43, 2015
|
|
4Z7Z
 
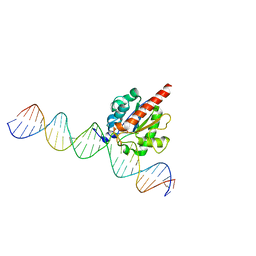 | | Structure of the enzyme-product complex resulting from TDG action on a GT mismatch in the presence of excess base | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, DNA (28-MER), ... | | Authors: | Pozharski, E, Malik, S.S, Drohat, A.C. | | Deposit date: | 2015-04-08 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Thymine DNA glycosylase exhibits negligible affinity for nucleobases that it removes from DNA.
Nucleic Acids Res., 43, 2015
|
|
4Z7B
 
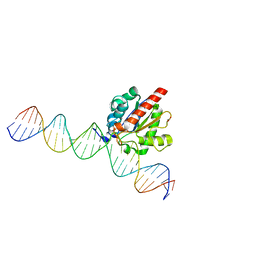 | | Structure of the enzyme-product complex resulting from TDG action on a GfC mismatch | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, DNA (28-MER), ... | | Authors: | Pozharski, E, Malik, S.S, Drohat, A.C. | | Deposit date: | 2015-04-07 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Thymine DNA glycosylase exhibits negligible affinity for nucleobases that it removes from DNA.
Nucleic Acids Res., 43, 2015
|
|
3CR2
 
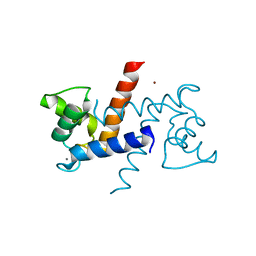 | | X-ray structure of bovine Zn(2+),Ca(2+)-S100B | | Descriptor: | CALCIUM ION, Protein S100-B, ZINC ION | | Authors: | Charpentier, T.H. | | Deposit date: | 2008-04-04 | | Release date: | 2008-06-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Divalent metal ion complexes of S100B in the absence and presence of pentamidine.
J.Mol.Biol., 382, 2008
|
|
3CR4
 
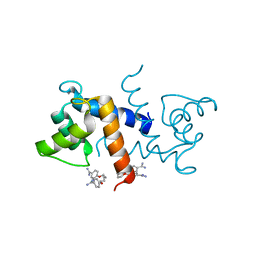 | | X-ray structure of bovine Pnt,Ca(2+)-S100B | | Descriptor: | 1,5-BIS(4-AMIDINOPHENOXY)PENTANE, CALCIUM ION, Protein S100-B | | Authors: | Charpentier, T.H. | | Deposit date: | 2008-04-04 | | Release date: | 2008-08-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Divalent metal ion complexes of S100B in the absence and presence of pentamidine.
J.Mol.Biol., 382, 2008
|
|
3CR5
 
 | | X-ray structure of bovine Pnt-Zn(2+),Ca(2+)-S100B | | Descriptor: | 1,5-BIS(4-AMIDINOPHENOXY)PENTANE, CALCIUM ION, Protein S100-B, ... | | Authors: | Charpentier, T.H. | | Deposit date: | 2008-04-04 | | Release date: | 2008-06-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Divalent metal ion complexes of S100B in the absence and presence of pentamidine.
J.Mol.Biol., 382, 2008
|
|
4Z3A
 
 | |
