5I93
 
 | | Structure of Mouse Acireductone dioxygenase with Ni2+ and 2-ketopentanoic acid in the active site | | Descriptor: | 1,2-dihydroxy-3-keto-5-methylthiopentene dioxygenase, 2-oxopentanoic acid, NICKEL (II) ION | | Authors: | Deshpande, A.R, Wagenpfeil, K, Pochapsky, T.C, Petsko, G.A, Ringe, D. | | Deposit date: | 2016-02-19 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.236 Å) | | Cite: | Metal-Dependent Function of a Mammalian Acireductone Dioxygenase.
Biochemistry, 55, 2016
|
|
6GCH
 
 | | STRUCTURE OF CHYMOTRYPSIN-*TRIFLUOROMETHYL KETONE INHIBITOR COMPLEXES. COMPARISON OF SLOWLY AND RAPIDLY EQUILIBRATING INHIBITORS | | Descriptor: | 1,1,1-TRIFLUORO-3-ACETAMIDO-4-PHENYL BUTAN-2-ONE(N-ACETYL-L-PHENYLALANYL TRIFLUOROMETHYL KETONE), GAMMA-CHYMOTRYPSIN A | | Authors: | Brady, K, Wei, A, Ringe, D, Abeles, R.H. | | Deposit date: | 1990-04-06 | | Release date: | 1990-10-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of chymotrypsin-trifluoromethyl ketone inhibitor complexes: comparison of slowly and rapidly equilibrating inhibitors.
Biochemistry, 29, 1990
|
|
1TPB
 
 | | OFFSET OF A CATALYTIC LESION BY A BOUND WATER SOLUBLE | | Descriptor: | PHOSPHOGLYCOLOHYDROXAMIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Zhang, Z, Sugio, S, Komives, E.A, Liu, K.D, Knowles, J.R, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-02-03 | | Release date: | 1995-02-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis for pseudoreversion of the E165D lesion by the secondary S96P mutation in triosephosphate isomerase depends on the positions of active site water molecules.
Biochemistry, 34, 1995
|
|
1TPC
 
 | | OFFSET OF A CATALYTIC LESION BY A BOUND WATER SOLUBLE | | Descriptor: | PHOSPHOGLYCOLOHYDROXAMIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Zhang, Z, Sugio, S, Komives, E.A, Liu, K.D, Knowles, J.R, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-02-03 | | Release date: | 1995-02-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis for pseudoreversion of the E165D lesion by the secondary S96P mutation in triosephosphate isomerase depends on the positions of active site water molecules.
Biochemistry, 34, 1995
|
|
1S5M
 
 | | Xylose Isomerase in Substrate and Inhibitor Michaelis States: Atomic Resolution Studies of a Metal-Mediated Hydride Shift | | Descriptor: | MANGANESE (II) ION, SODIUM ION, Xylose isomerase, ... | | Authors: | Fenn, T.D, Ringe, D, Petsko, G.A. | | Deposit date: | 2004-01-21 | | Release date: | 2004-02-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Xylose isomerase in substrate and inhibitor michaelis States: atomic resolution studies of a metal-mediated hydride shift(,).
Biochemistry, 43, 2004
|
|
1SCD
 
 | |
2INX
 
 | | Crystal Structure of Ketosteroid Isomerase D40N from Pseudomonas putida (pKSI) with bound 2,6-difluorophenol | | Descriptor: | 2,6-DIFLUOROPHENOL, Steroid delta-isomerase | | Authors: | Martinez Caaveiro, J.M, Pybus, B, Ringe, D, Petsko, G.A, Sigala, P, Kraut, D, Herschlag, D. | | Deposit date: | 2006-10-09 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Testing geometrical discrimination within an enzyme active site: constrained hydrogen bonding in the ketosteroid isomerase oxyanion hole.
J.Am.Chem.Soc., 130, 2008
|
|
1DAA
 
 | |
5DAA
 
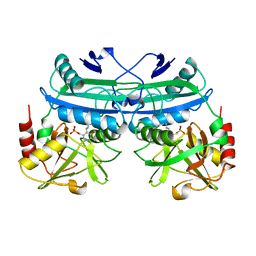 | |
1SCN
 
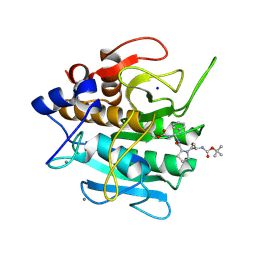 | | INACTIVATION OF SUBTILISIN CARLSBERG BY N-(TERT-BUTOXYCARBONYL-ALANYL-PROLYL-PHENYLALANYL)-O-BENZOL HYDROXYLAMINE: FORMATION OF COVALENT ENZYME-INHIBITOR LINKAGE IN THE FORM OF A CARBAMATE DERIVATIVE | | Descriptor: | CALCIUM ION, N-(tert-butoxycarbonyl)-L-alanyl-N-[(1R)-1-(carboxyamino)-2-phenylethyl]-L-prolinamide, SODIUM ION, ... | | Authors: | Steinmetz, A.C.U, Demuth, H.-U, Ringe, D. | | Deposit date: | 1994-03-02 | | Release date: | 1994-08-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Inactivation of subtilisin Carlsberg by N-((tert-butoxycarbonyl)alanylprolylphenylalanyl)-O-benzolhydroxyl- amine: formation of a covalent enzyme-inhibitor linkage in the form of a carbamate derivative.
Biochemistry, 33, 1994
|
|
1DDN
 
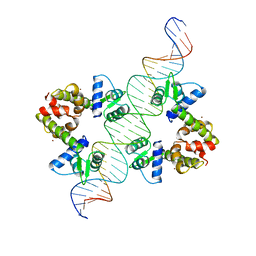 | | DIPHTHERIA TOX REPRESSOR (C102D MUTANT)/TOX DNA OPERATOR COMPLEX | | Descriptor: | 33 BASE DNA CONTAINING TOXIN OPERATOR, DIPHTHERIA TOX REPRESSOR, NICKEL (II) ION | | Authors: | White, A, Ding, X, Vanderspek, J.C, Murphy, J.R, Ringe, D. | | Deposit date: | 1998-06-23 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the metal-ion-activated diphtheria toxin repressor/tox operator complex.
Nature, 394, 1998
|
|
1DPR
 
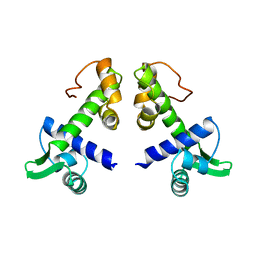 | | STRUCTURES OF THE APO-AND METAL ION ACTIVATED FORMS OF THE DIPHTHERIA TOX REPRESSOR FROM CORYNEBACTERIUM DIPHTHERIAE | | Descriptor: | DIPHTHERIA TOX REPRESSOR | | Authors: | Schiering, N, Tao, X, Murphy, J, Petsko, G.A, Ringe, D. | | Deposit date: | 1995-02-06 | | Release date: | 1995-09-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of the apo- and the metal ion-activated forms of the diphtheria tox repressor from Corynebacterium diphtheriae.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
5RHN
 
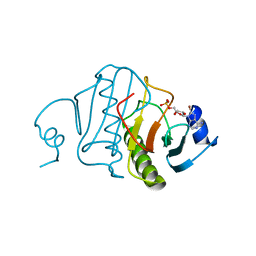 | | HISTIDINE TRIAD NUCLEOTIDE-BINDING PROTEIN (HINT) FROM RABBIT COMPLEXED WITH 8-BR-AMP | | Descriptor: | 8-BROMO-ADENOSINE-5'-MONOPHOSPHATE, HISTIDINE TRIAD NUCLEOTIDE-BINDING PROTEIN | | Authors: | Brenner, C, Garrison, P, Gilmour, J, Peisach, D, Ringe, D, Petsko, G.A, Lowenstein, J.M. | | Deposit date: | 1997-02-26 | | Release date: | 1997-06-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal structures of HINT demonstrate that histidine triad proteins are GalT-related nucleotide-binding proteins.
Nat.Struct.Biol., 4, 1997
|
|
1SCA
 
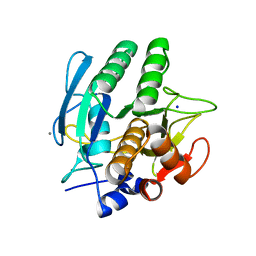 | | ENZYME CRYSTAL STRUCTURE IN A NEAT ORGANIC SOLVENT | | Descriptor: | CALCIUM ION, SODIUM ION, SUBTILISIN CARLSBERG | | Authors: | Fitzpatrick, P.A, Steinmetz, A.C.U, Ringe, D, Klibanov, A.M. | | Deposit date: | 1993-07-19 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Enzyme crystal structure in a neat organic solvent.
Proc.Natl.Acad.Sci.USA, 90, 1993
|
|
1RXT
 
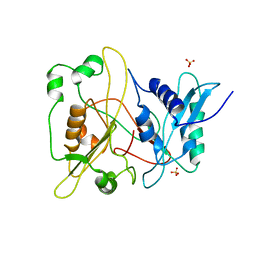 | | Crystal structure of human myristoyl-CoA:protein N-myristoyltransferase. | | Descriptor: | COBALT (II) ION, Glycylpeptide N-tetradecanoyltransferase 1, SULFATE ION | | Authors: | Yang, J, Wang, Y, Frey, G, Abeles, R.H, Petsko, G.A, Ringe, D. | | Deposit date: | 2003-12-18 | | Release date: | 2005-04-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of human myristoyl-CoA:protein N-myristoyltransferase
To be Published
|
|
1SCB
 
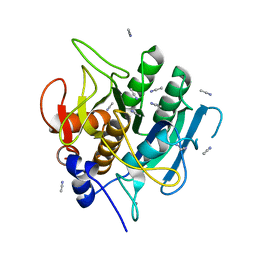 | | ENZYME CRYSTAL STRUCTURE IN A NEAT ORGANIC SOLVENT | | Descriptor: | ACETONITRILE, CALCIUM ION, SUBTILISIN CARLSBERG | | Authors: | Fitzpatrick, P.A, Steinmetz, A.C.U, Ringe, D, Klibanov, A.M. | | Deposit date: | 1993-07-13 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Enzyme crystal structure in a neat organic solvent.
Proc.Natl.Acad.Sci.USA, 90, 1993
|
|
1SFT
 
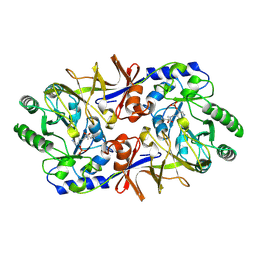 | | ALANINE RACEMASE | | Descriptor: | ACETATE ION, ALANINE RACEMASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Shaw, J.P, Petsko, G.A, Ringe, D. | | Deposit date: | 1996-09-20 | | Release date: | 1997-02-12 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Determination of the structure of alanine racemase from Bacillus stearothermophilus at 1.9-A resolution.
Biochemistry, 36, 1997
|
|
4DAA
 
 | |
5VWQ
 
 | | E.coli Aspartate aminotransferase-(1R,3S,4S)-3-amino-4-fluorocyclopentane-1-carboxylic acid (FCP) | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Aspartate aminotransferase | | Authors: | Mascarenhas, R, Lehrer, H, Liu, D, Ringe, D. | | Deposit date: | 2017-05-22 | | Release date: | 2017-08-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Selective Targeting by a Mechanism-Based Inactivator against Pyridoxal 5'-Phosphate-Dependent Enzymes: Mechanisms of Inactivation and Alternative Turnover.
Biochemistry, 56, 2017
|
|
3AAT
 
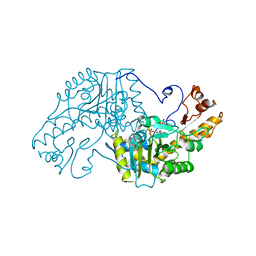 | |
5R42
 
 | | Crystal Structure of deuterated gamma-Chymotrypsin at pH 7.5, room temperature | | Descriptor: | IODIDE ION, gamma-Chymotrypsin, peptide SWPW, ... | | Authors: | Kreinbring, C.A, Wilson, M.A, Kovalevsky, A.Y, Blakeley, M.P, Fisher, S.Z, Lazar, L.M, Moulin, A.G, Novak, W.R, Petsko, G.A, Ringe, D. | | Deposit date: | 2020-02-18 | | Release date: | 2021-09-01 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Effect of Temperature and pH on Ionizable Residues in gamma-Chymotrypsin: a X-ray and Neutron Crystallography Study
To be published
|
|
5R49
 
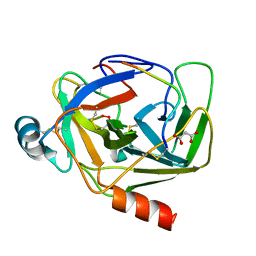 | | Crystal Structure of gamma-Chymotrypsin at pH 5.6, cryo temperature | | Descriptor: | IODIDE ION, MALONATE ION, gamma-chymotrypsin, ... | | Authors: | Kreinbring, C.A, Wilson, M.A, Kovalevsky, A.Y, Blakeley, M.P, Fisher, S.Z, Lazar, L.M, Moulin, A.G, Novak, W.R, Petsko, G.A, Ringe, D. | | Deposit date: | 2020-02-18 | | Release date: | 2021-09-01 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Effect of Temperature and pH on Ionizable Residues in gamma-Chymotrypsin: a X-ray and Neutron Crystallography Study
To be published
|
|
5R45
 
 | | Crystal Structure of gamma-Chymotrypsin at pH 7.5, cryo temperature | | Descriptor: | Chymotrypsinogen A, IODIDE ION, MALONATE ION, ... | | Authors: | Kreinbring, C.A, Wilson, M.A, Kovalevsky, A.Y, Blakeley, M.P, Fisher, S.Z, Lazar, L.M, Moulin, A.G, Novak, W.R, Petsko, G.A, Ringe, D. | | Deposit date: | 2020-02-18 | | Release date: | 2021-09-01 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Effect of Temperature and pH on Ionizable Residues in gamma-Chymotrypsin: a X-ray and Neutron Crystallography Study
To be published
|
|
5R4C
 
 | | Crystal Structure of gamma-Chymotrypsin at pH 9, room temperature | | Descriptor: | IODIDE ION, SULFATE ION, gamma-chymotrypsin, ... | | Authors: | Kreinbring, C.A, Wilson, M.A, Kovalevsky, A.Y, Blakeley, M.P, Fisher, S.Z, Lazar, L.M, Moulin, A.G, Novak, W.R, Petsko, G.A, Ringe, D. | | Deposit date: | 2020-02-18 | | Release date: | 2021-09-01 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Effect of Temperature and pH on Ionizable Residues in gamma-Chymotrypsin: a X-ray and Neutron Crystallography Study
To be published
|
|
5R48
 
 | | Crystal Structure of gamma-Chymotrypsin at pH 5.6, room temperature | | Descriptor: | IODIDE ION, SULFATE ION, gamma-chymotrypsin, ... | | Authors: | Kreinbring, C.A, Wilson, M.A, Kovalevsky, A.Y, Blakeley, M.P, Fisher, S.Z, Lazar, L.M, Moulin, A.G, Novak, W.R, Petsko, G.A, Ringe, D. | | Deposit date: | 2020-02-18 | | Release date: | 2021-09-01 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Effect of Temperature and pH on Ionizable Residues in gamma-Chymotrypsin: a X-ray and Neutron Crystallography Study
To be published
|
|
