2VP3
 
 | |
3C0W
 
 | | I-SceI in complex with a bottom nicked DNA substrate | | Descriptor: | CALCIUM ION, DNA (5'-D(*DAP*DCP*DGP*DCP*DTP*DAP*DGP*DGP*DGP*DAP*DTP*DAP*DAP*DCP*DAP*DGP*DGP*DGP*DTP*DAP*DAP*DTP*DAP*DC)-3'), DNA (5'-D(*DGP*DTP*DAP*DTP*DTP*DAP*DCP*DCP*DCP*DTP*DGP*DTP*DTP*DAP*DT)-3'), ... | | Authors: | Moure, C.M, Gimble, F.S, Quiocho, F.A. | | Deposit date: | 2008-01-21 | | Release date: | 2008-05-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of I-SceI complexed to nicked DNA substrates: snapshots of intermediates along the DNA cleavage reaction pathway.
Nucleic Acids Res., 36, 2008
|
|
3C0X
 
 | | I-SceI in complex with a top nicked DNA substrate | | Descriptor: | CALCIUM ION, DNA (5'-D(*DC*DAP*DCP*DGP*DCP*DTP*DAP*DGP*DGP*DGP*DAP*DTP*DAP*DA)-3'), DNA (5'-D(*DG*DGP*DTP*DAP*DTP*DTP*DAP*DCP*DCP*DCP*DTP*DGP*DTP*DTP*DAP*DTP*DCP*DCP*DCP*DTP*DAP*DGP*DCP*DGP*DT)-3'), ... | | Authors: | Moure, C.M, Gimble, F.S, Quiocho, F.A. | | Deposit date: | 2008-01-21 | | Release date: | 2008-05-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of I-SceI complexed to nicked DNA substrates: snapshots of intermediates along the DNA cleavage reaction pathway.
Nucleic Acids Res., 36, 2008
|
|
1GLG
 
 | |
2F3Z
 
 | | Calmodulin/IQ-AA domain complex | | Descriptor: | CALCIUM ION, Calmodulin, Voltage-dependent L-type calcium channel alpha-1C subunit | | Authors: | Fallon, J.L, Quiocho, F.A. | | Deposit date: | 2005-11-22 | | Release date: | 2005-12-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of Calmodulin Bound to the Hydrophobic IQ Domain of the Cardiac Ca(v)1.2 Calcium Channel.
Structure, 13, 2005
|
|
2F3Y
 
 | | Calmodulin/IQ domain complex | | Descriptor: | CALCIUM ION, Calmodulin, MAGNESIUM ION, ... | | Authors: | Fallon, J.L, Quiocho, F.A. | | Deposit date: | 2005-11-22 | | Release date: | 2005-12-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of Calmodulin Bound to the Hydrophobic IQ Domain of the Cardiac Ca(v)1.2 Calcium Channel.
Structure, 13, 2005
|
|
1M7I
 
 | | CRYSTAL STRUCTURE OF A MONOCLONAL FAB SPECIFIC FOR SHIGELLA FLEXNERI Y LIPOPOLYSACCHARIDE COMPLEXED WITH A PENTASACCHARIDE | | Descriptor: | alpha-L-rhamnopyranose-(1-2)-alpha-L-rhamnopyranose-(1-3)-alpha-L-rhamnopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-methyl 6-deoxy-alpha-L-rhamnopyranoside, heavy chain of the monoclonal antibody Fab SYA/J6, light chain of the monoclonal antibody Fab SYA/J6 | | Authors: | Vyas, N.K, Vyas, M.N, Chervenak, M.C, Johnson, M.A, Pinto, B.M, Bundle, D.R, Quiocho, F.A. | | Deposit date: | 2002-07-19 | | Release date: | 2003-07-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular Recognition of Oligosaccharide Epitopes by a Monoclonal Fab Specific for
Shigella flexneri Y Lipopolysaccharide: X-ray Structures and Thermodynamics
Biochemistry, 41, 2002
|
|
1MAR
 
 | |
3RRK
 
 | | Crystal structure of the cytoplasmic N-terminal domain of subunit I, homolog of subunit a, of V-ATPase | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, SULFATE ION, V-type ATPase 116 kDa subunit | | Authors: | Srinivasan, S, Vyas, N.K, Baker, M.L, Quiocho, F.A. | | Deposit date: | 2011-04-29 | | Release date: | 2011-08-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Crystal structure of the cytoplasmic N-terminal domain of subunit I, a homolog of subunit a, of V-ATPase.
J.Mol.Biol., 412, 2011
|
|
4W9N
 
 | | Enoyl-acyl carrier protein-reductase domain from human fatty acid synthase complexed with triclosan | | Descriptor: | CHLORIDE ION, Enoyl-[acyl-carrier-protein] reductase, IMIDAZOLE, ... | | Authors: | Sippel, K.H, Vyas, N.K, Sankaran, B, Quiocho, F.A. | | Deposit date: | 2014-08-27 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structure of the human Fatty Acid synthase enoyl-acyl carrier protein-reductase domain complexed with triclosan reveals allosteric protein-protein interface inhibition.
J.Biol.Chem., 289, 2014
|
|
2LIV
 
 | |
1SBP
 
 | |
1IXI
 
 | |
1IXG
 
 | |
1IXH
 
 | |
1UIO
 
 | | ADENOSINE DEAMINASE (HIS 238 ALA MUTANT) | | Descriptor: | 6-HYDROXY-7,8-DIHYDRO PURINE NUCLEOSIDE, ADENOSINE DEAMINASE, ZINC ION | | Authors: | Wilson, D.K, Quiocho, F.A. | | Deposit date: | 1996-08-30 | | Release date: | 1997-06-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Site-directed mutagenesis of histidine 238 in mouse adenosine deaminase: substitution of histidine 238 does not impede hydroxylate formation.
Biochemistry, 35, 1996
|
|
1UIP
 
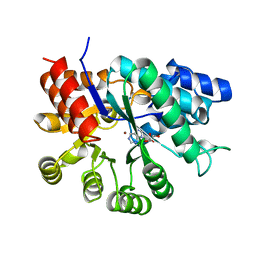 | | ADENOSINE DEAMINASE (HIS 238 GLU MUTANT) | | Descriptor: | ADENOSINE DEAMINASE, PURINE RIBOSIDE, ZINC ION | | Authors: | Wilson, D.K, Quiocho, F.A. | | Deposit date: | 1996-08-30 | | Release date: | 1997-06-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Site-directed mutagenesis of histidine 238 in mouse adenosine deaminase: substitution of histidine 238 does not impede hydroxylate formation.
Biochemistry, 35, 1996
|
|
1FKX
 
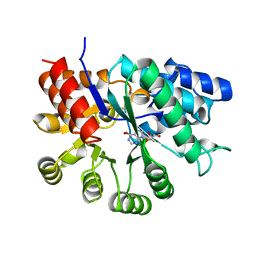 | | MURINE ADENOSINE DEAMINASE (D296A) | | Descriptor: | 6-HYDROXY-1,6-DIHYDRO PURINE NUCLEOSIDE, ADENOSINE DEAMINASE, ZINC ION | | Authors: | Wilson, D.K, Quiocho, F.A. | | Deposit date: | 1996-02-29 | | Release date: | 1996-08-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Probing the functional role of two conserved active site aspartates in mouse adenosine deaminase.
Biochemistry, 35, 1996
|
|
1FKW
 
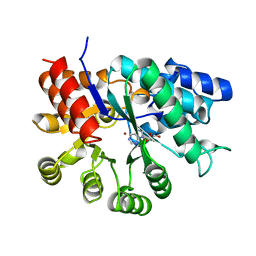 | | MURINE ADENOSINE DEAMINASE (D295E) | | Descriptor: | ADENOSINE DEAMINASE, PURINE RIBOSIDE, ZINC ION | | Authors: | Wilson, D.K, Quiocho, F.A. | | Deposit date: | 1996-02-29 | | Release date: | 1996-08-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Probing the functional role of two conserved active site aspartates in mouse adenosine deaminase.
Biochemistry, 35, 1996
|
|
2LBP
 
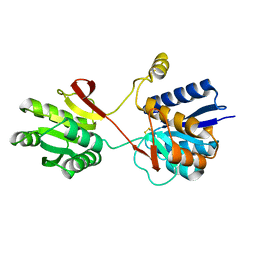 | |
2PFD
 
 | | Anisotropically refined structure of FTCD | | Descriptor: | Formimidoyltransferase-cyclodeaminase | | Authors: | Poon, B.K, Chen, X, Lu, M, Quiocho, F.A, Wang, Q, Ma, J. | | Deposit date: | 2007-04-04 | | Release date: | 2007-04-24 | | Last modified: | 2011-08-10 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Anisotropically refined structure of FTCD
To be Published
|
|
3G43
 
 | |
4CLN
 
 | | STRUCTURE OF A RECOMBINANT CALMODULIN FROM DROSOPHILA MELANOGASTER REFINED AT 2.2-ANGSTROMS RESOLUTION | | Descriptor: | CALCIUM ION, CALMODULIN | | Authors: | Taylor, D.A, Sack, J.S, Maune, J.F, Beckingham, K, Quiocho, F.A. | | Deposit date: | 1991-06-24 | | Release date: | 1992-07-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a recombinant calmodulin from Drosophila melanogaster refined at 2.2-A resolution.
J.Biol.Chem., 266, 1991
|
|
1APB
 
 | |
4DCG
 
 | | VACCINIA METHYLTRANSFERASE VP39 MUTANT D182A COMPLEXED WITH M7G AND S-ADENOSYLHOMOCYSTEINE | | Descriptor: | 7-METHYLGUANOSINE, S-ADENOSYL-L-HOMOCYSTEINE, VP39 | | Authors: | Hu, G, Hodel, A.E, Gershon, P.D, Quiocho, F.A. | | Deposit date: | 1998-07-13 | | Release date: | 1999-07-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | mRNA cap recognition: dominant role of enhanced stacking interactions between methylated bases and protein aromatic side chains.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
