3M0C
 
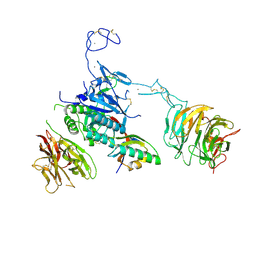 | |
4E9L
 
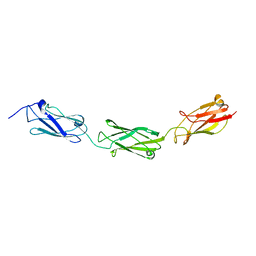 | | FdeC, a Novel Broadly Conserved Escherichia coli Adhesin Eliciting Protection against Urinary Tract Infections | | Descriptor: | Attaching and effacing protein, pathogenesis factor | | Authors: | Spraggon, G, Nesta, B, Alteri, C, Gomes Moriel, D, Rosini, R, Veggi, D, Smith, S, Bertoldi, I, Pastorello, I, Ferlenghi, I, Fontana, M.R, Frankel, G, Mobley, H.L.T, Rappuoli, R, Pizza, M, Serino, L, Soriana, M. | | Deposit date: | 2012-03-21 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | FdeC, a novel broadly conserved Escherichia coli adhesin eliciting protection against urinary tract infections.
MBio, 3, 2012
|
|
2HH5
 
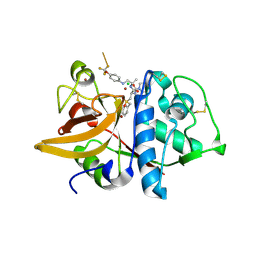 | | Crystal Structure of Cathepsin S in complex with a Zinc mediated non-covalent arylaminoethyl amide | | Descriptor: | CHLORIDE ION, Cathepsin S, N-[(1R)-1-[(BENZYLSULFONYL)METHYL]-2-{[(1S)-1-METHYL-2-{[4-(TRIFLUOROMETHOXY)PHENYL]AMINO}ETHYL]AMINO}-2-OXOETHYL]MORPHOLINE-4-CARBOXAMIDE, ... | | Authors: | Spraggon, G, Hornsby, M, Lesley, S.A, Tully, D.C, Harris, J.L, Karenewsky, D.S. | | Deposit date: | 2006-06-27 | | Release date: | 2006-08-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthesis and SAR of arylaminoethyl amides as noncovalent inhibitors of cathepsin S: P3 cyclic ethers.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
3E16
 
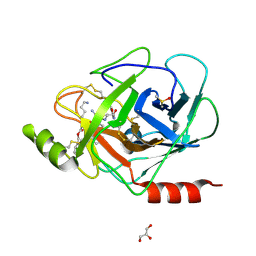 | | X-ray structure of human prostasin in complex with Benzoxazole warhead peptidomimic, lysine in P3 | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, Prostasin, ... | | Authors: | Spraggon, G, Hornsby, M, Shipway, A, Harris, J.L, Lesley, S.A. | | Deposit date: | 2008-08-01 | | Release date: | 2008-09-09 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of inhibitors of the channel-activating protease prostasin (CAP1/PRSS8) utilizing structure-based design.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3E0P
 
 | | The X-ray structure of Human Prostasin in complex with a covalent benzoxazole inhibitor | | Descriptor: | GLYCEROL, Prostasin, benzyl [(1R)-1-({(2S,4R)-2-({(1S)-5-amino-1-[(S)-1,3-benzoxazol-2-yl(hydroxy)methyl]pentyl}carbamoyl)-4-[(4-methylbenzyl)oxy]pyrrolidin-1-yl}carbonyl)-3-phenylpropyl]carbamate | | Authors: | Spraggon, G, Hornsby, M, Shipway, A, Harris, J.L, Lesley, S.A. | | Deposit date: | 2008-07-31 | | Release date: | 2008-09-09 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of inhibitors of the channel-activating protease prostasin (CAP1/PRSS8) utilizing structure-based design.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
4Z49
 
 | |
2OP3
 
 | | The structure of cathepsin S with a novel 2-arylphenoxyacetaldehyde inhibitor derived by the Substrate Activity Screening (SAS) method | | Descriptor: | 2,5,8,11,14,17,20,23,26,29,32,35,38,41,44,47,50,53,56,59,62,65,68,71,74,77,80-HEPTACOSAOXADOOCTACONTAN-82-OL, 2-[(2',3',4'-TRIFLUOROBIPHENYL-2-YL)OXY]ETHANOL, Cathepsin S, ... | | Authors: | Spraggon, G, Inagaki, H, Tsuruoka, H, Hornsby, M, Lesley, S.A, Ellman, J.A. | | Deposit date: | 2007-01-26 | | Release date: | 2007-05-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Characterization and optimization of selective, nonpeptidic inhibitors of cathepsin S with an unprecedented binding mode.
J.Med.Chem., 50, 2007
|
|
2F1G
 
 | | Cathepsin S in complex with non-covalent 2-(Benzoxazol-2-ylamino)-acetamide | | Descriptor: | Cathepsin S, GLYCEROL, N~2~-1,3-BENZOXAZOL-2-YL-3-CYCLOHEXYL-N-{2-[(4-METHOXYPHENYL)AMINO]ETHYL}-L-ALANINAMIDE | | Authors: | Spraggon, G, Hornsby, M, Lesley, S.A, Tully, D.C, Harris, J.L, Karenewsky, D.S, Kulathila, R, Clark, K. | | Deposit date: | 2005-11-14 | | Release date: | 2006-04-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Synthesis and evaluation of arylaminoethyl amides as noncovalent inhibitors of cathepsin S. Part 3: Heterocyclic P3.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2HHN
 
 | | Cathepsin S in complex with non covalent arylaminoethyl amide. | | Descriptor: | Cathepsin S, N-[(1R)-1-[(BENZYLSULFONYL)METHYL]-2-{[(1S)-1-METHYL-2-{[4-(TRIFLUOROMETHOXY)PHENYL]AMINO}ETHYL]AMINO}-2-OXOETHYL]MORPHOLINE-4-CARBOXAMIDE, SULFATE ION | | Authors: | Spraggon, G, Hornsby, M, Lesley, S.A, Tully, D.C, Harris, J.L, Karenewsky, D.S, Kulathila, R, Clark, K. | | Deposit date: | 2006-06-28 | | Release date: | 2007-05-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Synthesis and SAR of arylaminoethyl amides as noncovalent inhibitors of cathepsin S: P3 cyclic ethers
Bioorg.Med.Chem.Lett., 16, 2006
|
|
5LOZ
 
 | | STRUCTURE OF YEAST ENT1 ENTH DOMAIN | | Descriptor: | Epsin-1 | | Authors: | Tanner, N, Prag, G. | | Deposit date: | 2016-08-11 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A bacterial genetic selection system for ubiquitylation cascade discovery.
Nat.Methods, 13, 2016
|
|
5LN1
 
 | | STRUCTURE OF UBIQUITYLATED-RPN10 FROM YEAST; | | Descriptor: | 26S proteasome regulatory subunit RPN10, Polyubiquitin-B | | Authors: | Keren-Kaplan, T, Attali, I, Levin-Kravets, O, Prag, G. | | Deposit date: | 2016-08-02 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Structure of ubiquitylated-Rpn10 provides insight into its autoregulation mechanism.
Nat Commun, 7, 2016
|
|
4Z9S
 
 | |
5OJW
 
 | | S. cerevisiae UBC13 - MMs2 complex | | Descriptor: | Ubiquitin-conjugating enzyme E2 13, Ubiquitin-conjugating enzyme variant MMS2 | | Authors: | Sharon, I, Rathi, R, Levin-Kravets, O, Attali, I, Prag, G. | | Deposit date: | 2017-07-24 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | S. cerevisiae UBC13 - MMs2 complex
To Be Published
|
|
1FFR
 
 | | CRYSTAL STRUCTURE OF CHITINASE A MUTANT Y390F COMPLEXED WITH HEXA-N-ACETYLCHITOHEXAOSE (NAG)6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHITINASE A | | Authors: | Papanikolau, Y, Prag, G, Tavlas, G, Vorgias, C.E, Oppenheim, A.B, Petratos, K. | | Deposit date: | 2000-07-26 | | Release date: | 2001-09-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High resolution structural analyses of mutant chitinase A complexes with substrates provide new insight into the mechanism of catalysis.
Biochemistry, 40, 2001
|
|
5LP0
 
 | |
1U5T
 
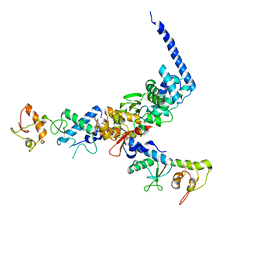 | | Structure of the ESCRT-II endosomal trafficking complex | | Descriptor: | Defective in vacuolar protein sorting; Vps36p, Hypothetical 23.6 kDa protein in YUH1-URA8 intergenic region, appears to be functionally related to SNF7; Snf8p | | Authors: | Hierro, A, Sun, J, Rusnak, A.S, Kim, J, Prag, G, Emr, S.D, Hurley, J.H. | | Deposit date: | 2004-07-28 | | Release date: | 2004-09-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure of ESCRT-II endosomal trafficking complex
Nature, 431, 2004
|
|
1EIB
 
 | | CRYSTAL STRUCTURE OF CHITINASE A MUTANT D313A COMPLEXED WITH OCTA-N-ACETYLCHITOOCTAOSE (NAG)8. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHITINASE A | | Authors: | Papanikolau, Y, Prag, G, Tavlas, G, Vorgias, C.E, Oppenheim, A.B, Petratos, K. | | Deposit date: | 2000-02-25 | | Release date: | 2001-02-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High resolution structural analyses of mutant chitinase A complexes with substrates provide new insight into the mechanism of catalysis.
Biochemistry, 40, 2001
|
|
1EHN
 
 | | CRYSTAL STRUCTURE OF CHITINASE A MUTANT E315Q COMPLEXED WITH OCTA-N-ACETYLCHITOOCTAOSE (NAG)8. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHITINASE A | | Authors: | Papanikolau, Y, Prag, G, Tavlas, G, Vorgias, C.E, Oppenheim, A.B, Petratos, K. | | Deposit date: | 2000-02-22 | | Release date: | 2001-02-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High resolution structural analyses of mutant chitinase A complexes with substrates provide new insight into the mechanism of catalysis.
Biochemistry, 40, 2001
|
|
6CUJ
 
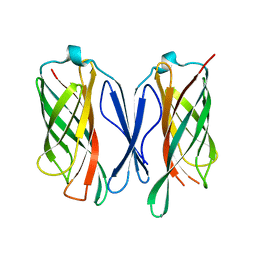 | |
7KFA
 
 | | PCSK9 in complex with PCSK9i a 13mer cyclic peptide LDLR disruptor | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, CALCIUM ION, Proprotein convertase subtilisin/kexin type 9, ... | | Authors: | Chopra, R, Xu, M, Spraggon, G. | | Deposit date: | 2020-10-13 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Identification of a PCSK9-LDLR disruptor peptide with in vivo function.
Cell Chem Biol, 29, 2022
|
|
6WYS
 
 | | Lon protease proteolytic domain | | Descriptor: | Lon protease homolog, mitochondrial, SULFATE ION | | Authors: | Lee, C.C, Spraggon, G. | | Deposit date: | 2020-05-13 | | Release date: | 2021-04-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.229 Å) | | Cite: | Structure-Based Design of Selective LONP1 Inhibitors for Probing In Vitro Biology.
J.Med.Chem., 64, 2021
|
|
6WZV
 
 | | Lon protease proteolytic domain | | Descriptor: | Lon protease homolog, mitochondrial, N-[(1R)-1-borono-3-methylbutyl]-Nalpha-(pyrazine-2-carbonyl)-D-phenylalaninamide, ... | | Authors: | Lee, C.C, Spraggon, G. | | Deposit date: | 2020-05-14 | | Release date: | 2021-04-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure-Based Design of Selective LONP1 Inhibitors for Probing In Vitro Biology.
J.Med.Chem., 64, 2021
|
|
6X27
 
 | |
6X1M
 
 | |
5W5O
 
 | |
