7LC7
 
 | |
4G7Y
 
 | |
7KWZ
 
 | | TDP-43 LCD amyloid fibrils | | Descriptor: | Isoform 2 of TAR DNA-binding protein 43 | | Authors: | Li, Q, Babinchak, W.M, Surewicz, W.K. | | Deposit date: | 2020-12-02 | | Release date: | 2021-02-24 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of amyloid fibrils formed by the entire low complexity domain of TDP-43.
Nat Commun, 12, 2021
|
|
4G80
 
 | |
4G7V
 
 | |
4HZY
 
 | | Crystal structure of influenza A neuraminidase N3-H274Y | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Neuraminidase | | Authors: | Li, Q, Qi, J, Vavricka, C.J, Gao, G.F. | | Deposit date: | 2012-11-16 | | Release date: | 2013-11-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | Functional and structural analysis of influenza virus neuraminidase N3 offers further insight into the mechanisms of oseltamivir resistance.
J.Virol., 87, 2013
|
|
4HZZ
 
 | | Crystal structure of influenza neuraminidase N3-H274Y complexed with oseltamivir | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Li, Q, Qi, J, Vavricka, C.J, Gao, G.F. | | Deposit date: | 2012-11-16 | | Release date: | 2013-11-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Functional and structural analysis of influenza virus neuraminidase N3 offers further insight into the mechanisms of oseltamivir resistance.
J.Virol., 87, 2013
|
|
4I00
 
 | | Crystal structure of influenza A neuraminidase N3-H274Y complexed with zanamivir | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Neuraminidase, ... | | Authors: | Li, Q, Qi, J, Vavricka, C.J, Gao, G.F. | | Deposit date: | 2012-11-16 | | Release date: | 2013-11-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Functional and structural analysis of influenza virus neuraminidase N3 offers further insight into the mechanisms of oseltamivir resistance.
J.Virol., 87, 2013
|
|
4HZV
 
 | | The crystal structure of influenza A neuraminidase N3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Li, Q, Qi, J, Vavricka, C.J, Gao, G.F. | | Deposit date: | 2012-11-15 | | Release date: | 2013-11-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Functional and structural analysis of influenza virus neuraminidase N3 offers further insight into the mechanisms of oseltamivir resistance.
J.Virol., 87, 2013
|
|
4HZW
 
 | | Crystal structure of influenza A neuraminidase N3 complexed with laninamivir | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 5-acetamido-2,6-anhydro-4-carbamimidamido-3,4,5-trideoxy-7-O-methyl-D-glycero-D-galacto-non-2-enonic acid, CALCIUM ION, ... | | Authors: | Li, Q, Qi, J, Vavricka, C.J, Gao, G.F. | | Deposit date: | 2012-11-15 | | Release date: | 2013-11-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Functional and structural analysis of influenza virus neuraminidase N3 offers further insight into the mechanisms of oseltamivir resistance.
J.Virol., 87, 2013
|
|
4HZX
 
 | | Crystal structure of influenza A neuraminidase N3 complexed with oseltamivir | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Li, Q, Qi, J, Vavricka, C.J, Gao, G.F. | | Deposit date: | 2012-11-15 | | Release date: | 2013-11-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Functional and structural analysis of influenza virus neuraminidase N3 offers further insight into the mechanisms of oseltamivir resistance.
J.Virol., 87, 2013
|
|
7W0V
 
 | | C4'-SCF3-DT modifeid DNA-DNA duplex | | Descriptor: | DNA (5'-D(*CP*CP*AP*TP*(DSW)P*AP*TP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*AP*TP*AP*AP*TP*GP*G)-3') | | Authors: | Li, Q, Trajkovski, M, Fan, C, Chen, J, Zhou, Y, Lu, K, Li, H, Su, X, Xi, Z, Plavec, J, Zhou, C. | | Deposit date: | 2021-11-18 | | Release date: | 2022-11-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 4'-SCF 3 -Labeling Constitutes a Sensitive 19 F NMR Probe for Characterization of Interactions in the Minor Groove of DNA.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
6ITH
 
 | |
8IOZ
 
 | | Crystal structure of transaminase | | Descriptor: | Branched chain amino acid: 2-keto-4-methylthiobutyrate aminotransferase | | Authors: | Li, Q, Zhu, Y.M, Gao, J, Wei, H.L, Han, X, Liu, W.D, Sun, Y.X. | | Deposit date: | 2023-03-13 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | structure of aminotransferase
To Be Published
|
|
8ISC
 
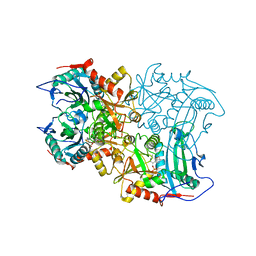 | | Crystal structure of MV in complex with LLP | | Descriptor: | Branched chain amino acid: 2-keto-4-methylthiobutyrate aminotransferase | | Authors: | Li, Q, Zhu, Y.M, Gao, J, Wei, H.L, Han, X, Liu, W.D, Sun, Y.X. | | Deposit date: | 2023-03-20 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structure of MV in complex with LLP
To Be Published
|
|
7BJI
 
 | | Crystal structure of the Danio rerio centrosomal protein Cep135 coiled-coil fragment 64-190 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Centrosomal protein of 135 kDa, ... | | Authors: | Li, Q, Hatzopoulos, G, Iller, O, Vakonakis, I. | | Deposit date: | 2021-01-14 | | Release date: | 2021-01-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Crystal structure of the Danio rerio centrosomal protein Cep135 coiled-coil fragment 64-190
To Be Published
|
|
8ZQA
 
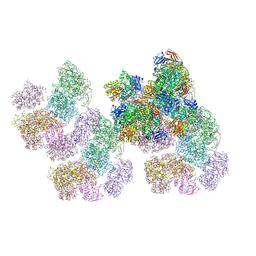 | |
8WJL
 
 | | Cryo-EM structure of 6-subunit Smc5/6 hinge region | | Descriptor: | E3 SUMO-protein ligase MMS21, Structural maintenance of chromosomes protein 5, Structural maintenance of chromosomes protein 6 | | Authors: | Li, Q, Zhang, J, Zhang, X, Cheng, T, Wang, Z, Jin, D, Chen, Z, Wang, L. | | Deposit date: | 2023-09-26 | | Release date: | 2024-06-26 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (6.15 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8WJN
 
 | | Cryo-EM structure of 6-subunit Smc5/6 head region | | Descriptor: | Non-structural maintenance of chromosome element 3, Non-structural maintenance of chromosomes element 1, Non-structural maintenance of chromosomes element 4, ... | | Authors: | Li, Q, Zhang, J, Zhang, X, Cheng, T, Wang, Z, Jin, D, Chen, Z, Wang, L. | | Deposit date: | 2023-09-26 | | Release date: | 2024-06-26 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (5.58 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8WJO
 
 | | Cryo-EM structure of 8-subunit Smc5/6 arm region | | Descriptor: | DNA repair protein KRE29, E3 SUMO-protein ligase MMS21, Structural maintenance of chromosomes protein 5, ... | | Authors: | Li, Q, Zhang, J, Zhang, X, Cheng, T, Wang, Z, Jin, D, Chen, Z, Wang, L. | | Deposit date: | 2023-09-26 | | Release date: | 2024-06-26 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (6.04 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7RK0
 
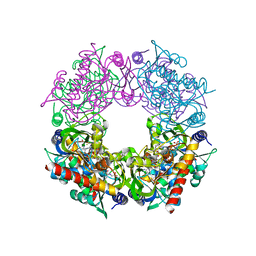 | | Crystal structure of Thermovibrio ammonificans THI4 | | Descriptor: | 2-[(E)-[(4R)-5-[[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-4-oxidanyl-3-oxidanylidene-pentan-2-ylidene]amino]ethanoic acid, FE (III) ION, Thiamine thiazole synthase | | Authors: | Li, Q, Bruner, S.D. | | Deposit date: | 2021-07-21 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structure and function of aerotolerant, multiple-turnover THI4 thiazole synthases.
Biochem.J., 478, 2021
|
|
2L4R
 
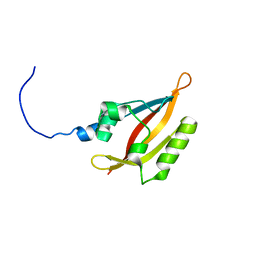 | | NMR solution structure of the N-terminal PAS domain of hERG | | Descriptor: | Potassium voltage-gated channel subfamily H member 2 | | Authors: | Gayen, N, Li, Q, Chen, A.S, Huang, Q, Raida, M, Kang, C. | | Deposit date: | 2010-10-13 | | Release date: | 2010-12-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the N-terminal domain of hERG and its interaction with the S4-S5 linker.
Biochem.Biophys.Res.Commun., 403, 2010
|
|
5TX3
 
 | | Structure of Maternal Embryonic Leucine Zipper Kinase | | Descriptor: | 7-[(1S)-4-hydroxy-2,3-dihydro-1H-inden-1-yl]-5,5-dimethyl-2-({3-[(pyrrolidin-1-yl)methyl]phenyl}amino)-5,7-dihydro-6H-pyrrolo[2,3-d]pyrimidin-6-one, Maternal embryonic leucine zipper kinase | | Authors: | Li, Q, Seo, H.-S, Huang, H.-T, Gray, N.S, Dhe-Paganon, S, Eck, M.J. | | Deposit date: | 2016-11-15 | | Release date: | 2017-11-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | MELK is not necessary for the proliferation of basal-like breast cancer cells.
Elife, 6, 2017
|
|
5TWU
 
 | | Structure of Maternal Embryonic Leucine Zipper Kinase | | Descriptor: | Maternal embryonic leucine zipper kinase | | Authors: | Li, Q, Seo, H.-S, Huang, H.-T, Gray, N.S, Dhe-Paganon, S, Eck, M.J. | | Deposit date: | 2016-11-14 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | MELK is not necessary for the proliferation of basal-like breast cancer cells.
Elife, 6, 2017
|
|
6D2I
 
 | | JAK2 Pseudokinase V617F in complex with AT9283 | | Descriptor: | 1-cyclopropyl-3-{3-[5-(morpholin-4-ylmethyl)-1H-benzimidazol-2-yl]-1H-pyrazol-4-yl}urea, Tyrosine-protein kinase | | Authors: | Li, Q, Li, K, Eck, M.J. | | Deposit date: | 2018-04-13 | | Release date: | 2019-03-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.192 Å) | | Cite: | Discovery and Structural Characterization of ATP-Site Ligands for the Wild-Type and V617F Mutant JAK2 Pseudokinase Domain.
ACS Chem. Biol., 14, 2019
|
|
