3X00
 
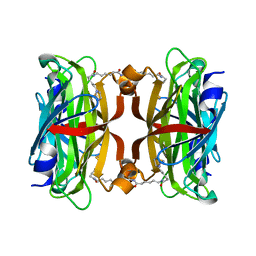 | | Crystal structure of the core streptavidin mutant V212 (Y22S/N23D/S27D/S45N/Y83S/R84K/E101D/R103K/E116N) complexed with bis iminobiotin long tail (Bis-IMNtail) at 1.3 A resolution | | Descriptor: | 6-({5-[(2E,3aS,4S,6aR)-2-iminohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoyl}amino)hexanoic acid, ETHANE-1,2-DIAMINE, Streptavidin | | Authors: | Kawato, T, Mizohata, E, Shimizu, Y, Meshizuka, T, Yamamoto, T, Takasu, N, Matsuoka, M, Matsumura, H, Kodama, T, Kanai, M, Doi, H, Inoue, T, Sugiyama, A. | | Deposit date: | 2014-10-09 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure-based design and synthesis of a bivalent iminobiotin analog showing strong affinity toward a low immunogenic streptavidin mutant.
Biosci.Biotechnol.Biochem., 79, 2015
|
|
1AYG
 
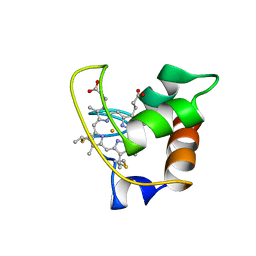 | | SOLUTION STRUCTURE OF CYTOCHROME C-552, NMR, 20 STRUCTURES | | Descriptor: | CYTOCHROME C-552, HEME C | | Authors: | Hasegawa, J, Yoshida, T, Yamazaki, T, Sambongi, Y, Yu, Y, Igarashi, Y, Kodama, T, Yamazaki, K, Hakusui, H, Kyogoku, Y, Kobayashi, Y. | | Deposit date: | 1997-11-04 | | Release date: | 1998-11-25 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of thermostable cytochrome c-552 from Hydrogenobacter thermophilus determined by 1H-NMR spectroscopy.
Biochemistry, 37, 1998
|
|
6A78
 
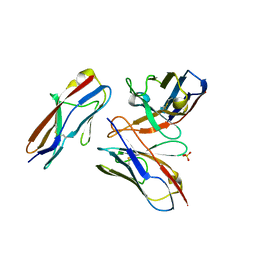 | | Crystal structure of the fifth immunoglobulin domain (Ig5) of human Robo1 in complex with the scFv fragment of murine monoclonal antibody B5209B | | Descriptor: | Heavy chain and linker region of the anti-human Robo1 antibody B5209B scFv, Light chain region of the anti-human Robo1 antibody B5209B scFv, Roundabout homolog 1, ... | | Authors: | Mizohata, E, Nakayama, T, Kado, Y, Inoue, T. | | Deposit date: | 2018-07-02 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Affinity Improvement of a Cancer-Targeted Antibody through Alanine-Induced Adjustment of Antigen-Antibody Interface.
Structure, 27, 2019
|
|
6A77
 
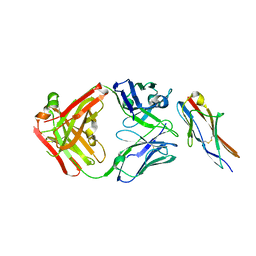 | | Crystal structure of the fifth immunoglobulin domain (Ig5) of human Robo1 in complex with the Fab fragment of murine monoclonal antibody B5209B | | Descriptor: | Heavy chain of the anti-human Robo1 antibody B5209B Fab, Light chain of the anti-human Robo1 antibody B5209B Fab, Roundabout homolog 1 | | Authors: | Mizohata, E, Nakayama, T, Kado, Y, Inoue, T. | | Deposit date: | 2018-07-02 | | Release date: | 2019-01-30 | | Last modified: | 2019-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Affinity Improvement of a Cancer-Targeted Antibody through Alanine-Induced Adjustment of Antigen-Antibody Interface.
Structure, 27, 2019
|
|
6A76
 
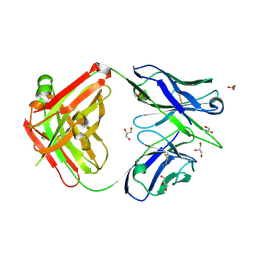 | | Crystal structure of the Fab fragment of B5209B, a murine monoclonal antibody specific for the fifth immunoglobulin domain (Ig5) of human ROBO1 | | Descriptor: | GLYCEROL, Heavy chain of the anti-human Robo1 antibody B5209B Fab, Light chain of the anti-human Robo1 antibody B5209B Fab, ... | | Authors: | Mizohata, E, Nakayama, T, Kado, Y, Inoue, T. | | Deposit date: | 2018-07-02 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Affinity Improvement of a Cancer-Targeted Antibody through Alanine-Induced Adjustment of Antigen-Antibody Interface.
Structure, 27, 2019
|
|
6A79
 
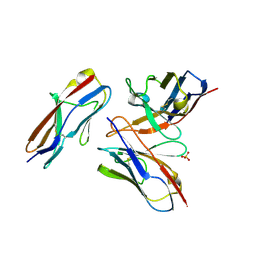 | | Crystal structure of the fifth immunoglobulin domain (Ig5) of human Robo1 in complex with the mutant scFv fragment (P103A) of murine monoclonal antibody B5209B | | Descriptor: | Heavy chain of the anti-human Robo1 antibody B5209B scFv, Light chain region of the anti-human Robo1 antibody B5209B scFv, Roundabout homolog 1, ... | | Authors: | Mizohata, E, Nakayama, T, Kado, Y, Yokota, Y, Inoue, T. | | Deposit date: | 2018-07-02 | | Release date: | 2019-01-30 | | Last modified: | 2019-03-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Affinity Improvement of a Cancer-Targeted Antibody through Alanine-Induced Adjustment of Antigen-Antibody Interface.
Structure, 27, 2019
|
|
7VI4
 
 | | Electron crystallographic structure of TIA-1 prion-like domain, A381T mutant | | Descriptor: | TIA-1 prion-like domain | | Authors: | Takaba, K, Maki-Yonekura, S, Sekiyama, N, Imamura, K, Kodama, T, Tochio, H, Yonekura, K. | | Deposit date: | 2021-09-24 | | Release date: | 2022-09-28 | | Last modified: | 2024-06-19 | | Method: | ELECTRON CRYSTALLOGRAPHY (0.95 Å) | | Cite: | ALS mutations in the TIA-1 prion-like domain trigger highly condensed pathogenic structures.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7VI5
 
 | | Electron crystallographic structure of TIA-1 prion-like domain, wild type sequence | | Descriptor: | TIA-1 prion-like domain | | Authors: | Takaba, K, Maki-Yonekura, S, Sekiyama, N, Imamura, K, Kodama, T, Tochio, H, Yonekura, K. | | Deposit date: | 2021-09-24 | | Release date: | 2022-09-28 | | Last modified: | 2023-11-29 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.761 Å) | | Cite: | ALS mutations in the TIA-1 prion-like domain trigger highly condensed pathogenic structures.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
1YNR
 
 | | Crystal structure of the cytochrome c-552 from Hydrogenobacter thermophilus at 2.0 resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cytochrome c-552, HEME C, ... | | Authors: | Travaglini-Allocatelli, C, Gianni, S, Dubey, V.K, Borgia, A, Di Matteo, A, Bonivento, D, Cutruzzola, F, Bren, K.L, Brunori, M. | | Deposit date: | 2005-01-25 | | Release date: | 2005-05-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An Obligatory Intermediate in the Folding Pathway of Cytochrome c552 from Hydrogenobacter thermophilus
J.Biol.Chem., 280, 2005
|
|
5B0D
 
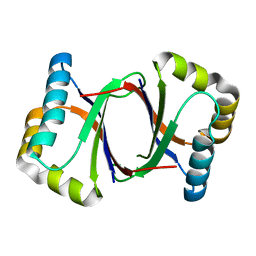 | | Polyketide cyclase OAC from Cannabis sativa, Y27W mutant | | Descriptor: | Olivetolic acid cyclase | | Authors: | Yang, X, Matsui, T, Mori, T, Abe, I, Morita, H. | | Deposit date: | 2015-10-28 | | Release date: | 2016-01-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structural basis for olivetolic acid formation by a polyketide cyclase from Cannabis sativa
Febs J., 283, 2016
|
|
5B0C
 
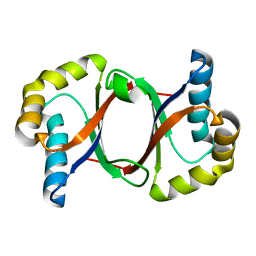 | | Polyketide cyclase OAC from Cannabis sativa, Y27F mutant | | Descriptor: | Olivetolic acid cyclase | | Authors: | Yang, X, Matsui, T, Mori, T, Abe, I, Morita, H. | | Deposit date: | 2015-10-28 | | Release date: | 2016-01-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Structural basis for olivetolic acid formation by a polyketide cyclase from Cannabis sativa
Febs J., 283, 2016
|
|
5B0B
 
 | | Polyketide cyclase OAC from Cannabis sativa, I7F mutant | | Descriptor: | ACETATE ION, Olivetolic acid cyclase | | Authors: | Yang, X, Matsui, T, Mori, T, Abe, I, Morita, H. | | Deposit date: | 2015-10-28 | | Release date: | 2016-01-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.187 Å) | | Cite: | Structural basis for olivetolic acid formation by a polyketide cyclase from Cannabis sativa
Febs J., 283, 2016
|
|
5B09
 
 | | Polyketide cyclase OAC from Cannabis sativa bound with Olivetolic acid | | Descriptor: | 2,4-bis(oxidanyl)-6-pentyl-benzoic acid, Olivetolic acid cyclase | | Authors: | Yang, X, Matsui, T, Mori, T, Abe, I, Morita, H. | | Deposit date: | 2015-10-28 | | Release date: | 2016-01-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for olivetolic acid formation by a polyketide cyclase from Cannabis sativa
Febs J., 283, 2016
|
|
5B0G
 
 | | Polyketide cyclase OAC from Cannabis sativa, H78S mutant | | Descriptor: | Olivetolic acid cyclase | | Authors: | Yang, X, Matsui, T, Mori, T, Abe, I, Morita, H. | | Deposit date: | 2015-10-28 | | Release date: | 2016-01-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for olivetolic acid formation by a polyketide cyclase from Cannabis sativa
Febs J., 283, 2016
|
|
5B08
 
 | | Polyketide cyclase OAC from Cannabis sativa | | Descriptor: | Olivetolic acid cyclase | | Authors: | Yang, X, Matsui, T, Mori, T, Abe, I, Morita, H. | | Deposit date: | 2015-10-28 | | Release date: | 2016-01-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.325 Å) | | Cite: | Structural basis for olivetolic acid formation by a polyketide cyclase from Cannabis sativa
Febs J., 283, 2016
|
|
5B0A
 
 | | Polyketide cyclase OAC from Cannabis sativa, H5Q mutant | | Descriptor: | Olivetolic acid cyclase | | Authors: | Yang, X, Matsui, T, Mori, T, Abe, I, Morita, H. | | Deposit date: | 2015-10-28 | | Release date: | 2016-01-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for olivetolic acid formation by a polyketide cyclase from Cannabis sativa
Febs J., 283, 2016
|
|
5B0E
 
 | | Polyketide cyclase OAC from Cannabis sativa, V59M mutant | | Descriptor: | GLYCEROL, Olivetolic acid cyclase | | Authors: | Yang, X, Matsui, T, Mori, T, Abe, I, Morita, H. | | Deposit date: | 2015-10-28 | | Release date: | 2016-01-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.603 Å) | | Cite: | Structural basis for olivetolic acid formation by a polyketide cyclase from Cannabis sativa
Febs J., 283, 2016
|
|
5B0F
 
 | | Polyketide cyclase OAC from Cannabis sativa, Y72F mutant | | Descriptor: | GLYCEROL, Olivetolic acid cyclase | | Authors: | Yang, X, Matsui, T, Mori, T, Abe, I, Morita, H. | | Deposit date: | 2015-10-28 | | Release date: | 2016-01-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for olivetolic acid formation by a polyketide cyclase from Cannabis sativa
Febs J., 283, 2016
|
|
3AOX
 
 | | X-ray crystal structure of human anaplastic lymphoma kinase in complex with CH5424802 | | Descriptor: | 1,2-ETHANEDIOL, 9-ethyl-6,6-dimethyl-8-[4-(morpholin-4-yl)piperidin-1-yl]-11-oxo-6,11-dihydro-5H-benzo[b]carbazole-3-carbonitrile, ALK tyrosine kinase receptor | | Authors: | Nagel, S, Moertl, M, Jestel, A, Fukami, T.A. | | Deposit date: | 2010-10-08 | | Release date: | 2011-05-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | CH5424802, a selective ALK inhibitor capable of blocking the resistant gatekeeper mutant
Cancer Cell, 19, 2011
|
|
7FFH
 
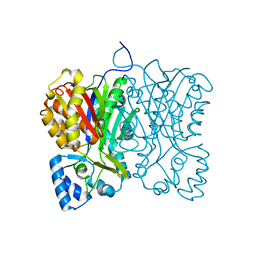 | | Diarylpentanoid-producing polyketide synthase (N199L mutant) | | Descriptor: | Type III polyketide synthase | | Authors: | Morita, H, Wong, C.P, Liu, Q, Takeshi, K, Lee, Y, Nakashima, Y. | | Deposit date: | 2021-07-23 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of a diarylpentanoid-producing polyketide synthase revealing an unusual biosynthetic pathway of 2-(2-phenylethyl)chromones in agarwood.
Nat Commun, 13, 2022
|
|
7XVJ
 
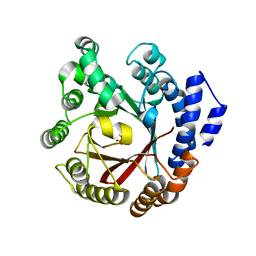 | | Crystal structure of CdpNPT in complex with harmol | | Descriptor: | 1-methyl-9~{H}-pyrido[3,4-b]indol-7-ol, Cyclic dipeptide N-prenyltransferase, PHOSPHATE ION | | Authors: | Nakashima, Y, Morita, H. | | Deposit date: | 2022-05-24 | | Release date: | 2023-04-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Enzymatic formation of a prenyl beta-carboline by a fungal indole prenyltransferase.
J Nat Med, 76, 2022
|
|
8XDU
 
 | |
8XE4
 
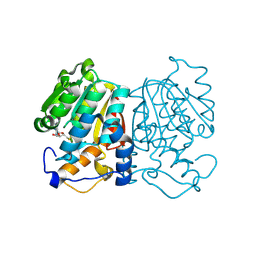 | | norbelladine 4'-O-methyltransferase complexed with Mg, SAH, and norbelladine | | Descriptor: | GLYCEROL, MAGNESIUM ION, Norbelladine, ... | | Authors: | Saw, Y.Y.H, Nakashima, Y, Morita, H. | | Deposit date: | 2023-12-11 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure-Based Catalytic Mechanism of Amaryllidaceae O-Methyltransferases
Acs Catalysis, 2024
|
|
8XDP
 
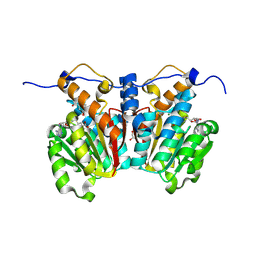 | | O-methyltransferase from Lycoris longituba complexed with Mg, SAH, and 3,4-dihydroxybenzaldehyde | | Descriptor: | GLYCEROL, MAGNESIUM ION, Protocatechuic aldehyde, ... | | Authors: | Saw, Y.Y.H, Nakashima, Y, Morita, H. | | Deposit date: | 2023-12-11 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-Based Catalytic Mechanism of Amaryllidaceae O-Methyltransferases
Acs Catalysis, 2024
|
|
8XDQ
 
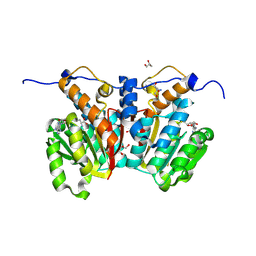 | | O-methyltransferase from Lycoris aurea complexed with Mg and SAH | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Saw, Y.Y.H, Nakashima, Y, Morita, H. | | Deposit date: | 2023-12-11 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structure-Based Catalytic Mechanism of Amaryllidaceae O-Methyltransferases
Acs Catalysis, 2024
|
|
