8E94
 
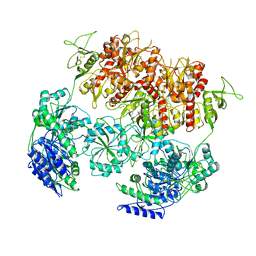 | | PYD-106-bound Human GluN1a-GluN2C NMDA receptor in intact conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chou, T.-H, Furukawa, H. | | Deposit date: | 2022-08-26 | | Release date: | 2022-12-07 | | Last modified: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Structural insights into assembly and function of GluN1-2C, GluN1-2A-2C, and GluN1-2D NMDARs.
Mol.Cell, 82, 2022
|
|
8E93
 
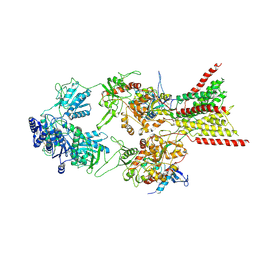 | | D-cycloserine and glutamate bound Human GluN1a-GluN2C NMDA receptor in splayed conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | Authors: | Chou, T.-H, Furukawa, H. | | Deposit date: | 2022-08-26 | | Release date: | 2022-12-07 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Structural insights into assembly and function of GluN1-2C, GluN1-2A-2C, and GluN1-2D NMDARs.
Mol.Cell, 82, 2022
|
|
8E99
 
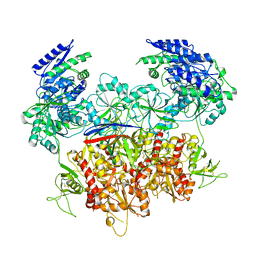 | | Human GluN1a-GluN2A-GluN2C triheteromeric NMDA receptor in complex with Nb-4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | Authors: | Chou, T.-H, Furukawa, H. | | Deposit date: | 2022-08-26 | | Release date: | 2022-12-07 | | Last modified: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (4.24 Å) | | Cite: | Structural insights into assembly and function of GluN1-2C, GluN1-2A-2C, and GluN1-2D NMDARs.
Mol.Cell, 82, 2022
|
|
8E92
 
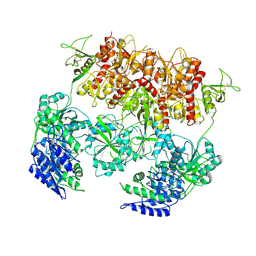 | | D-cycloserine and glutamate bound Human GluN1a-GluN2C NMDA receptor in intact conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | Authors: | Chou, T.-H, Furukawa, H. | | Deposit date: | 2022-08-26 | | Release date: | 2022-12-07 | | Last modified: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Structural insights into assembly and function of GluN1-2C, GluN1-2A-2C, and GluN1-2D NMDARs.
Mol.Cell, 82, 2022
|
|
8E98
 
 | | D-cycloserine and glutamate bound Human GluN1a-GluN2C NMDA receptor in nanodisc - intact conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, NMDA 1, ... | | Authors: | Chou, T.-H, Furukawa, H. | | Deposit date: | 2022-08-26 | | Release date: | 2022-12-07 | | Last modified: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Structural insights into assembly and function of GluN1-2C, GluN1-2A-2C, and GluN1-2D NMDARs.
Mol.Cell, 82, 2022
|
|
8J46
 
 | | Human Consensus Olfactory Receptor OR52c in apo state, OR52c-bRIL | | Descriptor: | Olfactory receptor OR52c,Soluble cytochrome b562 | | Authors: | Choi, C.W, Bae, J, Choi, H.-J, Kim, J. | | Deposit date: | 2023-04-19 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Understanding the molecular mechanisms of odorant binding and activation of the human OR52 family.
Nat Commun, 14, 2023
|
|
2M09
 
 | | Structure, phosphorylation and U2AF65 binding of the Nterminal Domain of splicing factor 1 during 3 splice site Recognition | | Descriptor: | Splicing factor 1 | | Authors: | Madl, T, Sattler, M, Zhang, Y, Bagdiul, I, Kern, T, Kang, H, Zou, P, Maeusbacher, N, Sieber, S.A, Kraemer, A. | | Deposit date: | 2012-10-22 | | Release date: | 2013-01-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure, phosphorylation and U2AF65 binding of the N-terminal domain of splicing factor 1 during 3'-splice site recognition.
Nucleic Acids Res., 41, 2013
|
|
2M0G
 
 | | Structure, phosphorylation and U2AF65 binding of the Nterminal Domain of splicing factor 1 during 3 splice site Recognition | | Descriptor: | Splicing factor 1, Splicing factor U2AF 65 kDa subunit | | Authors: | Madl, T, Sattler, M, Zhang, Y, Bagdiul, I, Kern, T, Kang, H, Zou, P, Maeusbacher, N, Sieber, S.A, Kraemer, A. | | Deposit date: | 2012-10-25 | | Release date: | 2013-01-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure, phosphorylation and U2AF65 binding of the N-terminal domain of splicing factor 1 during 3'-splice site recognition.
Nucleic Acids Res., 41, 2013
|
|
2KMD
 
 | | Ras signaling requires dynamic properties of Ets1 for phosphorylation-enhanced binding to co-activator CBP | | Descriptor: | Protein C-ets-1 | | Authors: | Nelson, M.L, Kang, H, Lee, G.M, Blaszczak, A.G, Lau, D.K.W, McIntosh, L.P, Graves, B.J. | | Deposit date: | 2009-07-27 | | Release date: | 2010-05-05 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Ras signaling requires dynamic properties of Ets1 for phosphorylation-enhanced binding to coactivator CBP.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2KXX
 
 | | NMR Structure of Escherichia coli BamE, a Lipoprotein Component of the beta-Barrel Assembly Machinery Complex | | Descriptor: | Small protein A | | Authors: | Kim, K, Okon, M, Escobar, E, Kang, H, McIntosh, L, Paetzel, M. | | Deposit date: | 2010-05-13 | | Release date: | 2011-01-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of Escherichia coli BamE, a Lipoprotein Component of the beta-Barrel Assembly Machinery Complex.
Biochemistry, 50, 2011
|
|
8KGN
 
 | | Structure of African swine fever virus topoisomerase II in complex with dsDNA | | Descriptor: | DNA (38-MER), DNA topoisomerase 2 | | Authors: | Cong, J, Xin, Y, Li, X, Chen, Y. | | Deposit date: | 2023-08-19 | | Release date: | 2024-04-03 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Structural insights into the DNA topoisomerase II of the African swine fever virus.
Nat Commun, 15, 2024
|
|
8KGT
 
 | | Structure of African swine fever virus topoisomerase II in complex with dsDNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA topoisomerase 2 | | Authors: | Cong, J, Xin, Y, Li, X, Chen, Y. | | Deposit date: | 2023-08-19 | | Release date: | 2024-04-03 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the DNA topoisomerase II of the African swine fever virus.
Nat Commun, 15, 2024
|
|
8KGS
 
 | | Structure of African swine fever virus topoisomerase II in complex with dsDNA | | Descriptor: | DNA topoisomerase 2, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Cong, J, Xin, Y, Li, X, Chen, Y. | | Deposit date: | 2023-08-19 | | Release date: | 2024-04-03 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Structural insights into the DNA topoisomerase II of the African swine fever virus.
Nat Commun, 15, 2024
|
|
8KGL
 
 | |
8KGO
 
 | |
8KGR
 
 | | Structure of African swine fever virus topoisomerase II in complex with dsDNA | | Descriptor: | DNA (32-MER), DNA (33-MER), DNA topoisomerase 2, ... | | Authors: | Cong, J, Xin, U, Li, X, Chen, Y. | | Deposit date: | 2023-08-19 | | Release date: | 2024-04-03 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into the DNA topoisomerase II of the African swine fever virus.
Nat Commun, 15, 2024
|
|
8KGM
 
 | | Structure of African swine fever virus topoisomerase II in complex with dsDNA | | Descriptor: | DNA (38-MER), DNA topoisomerase 2 | | Authors: | Cong, J, Xin, Y, Li, X, Chen, Y. | | Deposit date: | 2023-08-19 | | Release date: | 2024-04-03 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural insights into the DNA topoisomerase II of the African swine fever virus.
Nat Commun, 15, 2024
|
|
8KGP
 
 | |
8KGQ
 
 | | Structure of African swine fever virus topoisomerase II in complex with dsDNA | | Descriptor: | DNA (38-MER), DNA topoisomerase 2 | | Authors: | Cong, J, Xin, Y, Li, X, Chen, Y. | | Deposit date: | 2023-08-19 | | Release date: | 2024-04-03 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Structural insights into the DNA topoisomerase II of the African swine fever virus.
Nat Commun, 15, 2024
|
|
7JX4
 
 | | Crystal Structure of N-Lysine Peptoid-modified Collagen Triple Helix | | Descriptor: | Collagen mimetic peptide with N-Lysine guest | | Authors: | Yu, M.S, Whitby, F.G, Hill, C.P, Kessler, J.L, Yang, L.D. | | Deposit date: | 2020-08-26 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Peptoid Residues Make Diverse, Hyperstable Collagen Triple-Helices.
J.Am.Chem.Soc., 143, 2021
|
|
7JX5
 
 | | Crystal Structure of N-Phenylalanine Peptoid-modified Collagen Triple Helix | | Descriptor: | Collagen mimetic peptide with N-Phenylalanine guest | | Authors: | Yu, M.S, Whitby, F.G, Hill, C.P, Kessler, J.L, Yang, L.D. | | Deposit date: | 2020-08-26 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Peptoid Residues Make Diverse, Hyperstable Collagen Triple-Helices
J.Am.Chem.Soc., 143, 2021
|
|
7YOV
 
 | |
7YOT
 
 | |
7YOU
 
 | |
8W77
 
 | | Human Consensus Olfactory Receptor OR52c in apo state, OR52c only | | Descriptor: | Human Consensus Olfactory Receptor OR52c in apo state, receptor only,Soluble cytochrome b562 | | Authors: | Choi, C.W, Bae, J, Choi, H.-J, Kim, J. | | Deposit date: | 2023-08-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Understanding the molecular mechanisms of odorant binding and activation of the human OR52 family.
Nat Commun, 14, 2023
|
|
