4OCX
 
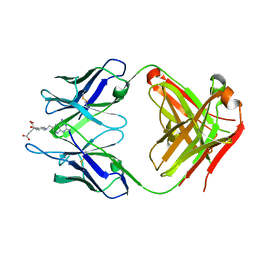 | | Fab complex with methotrexate | | Descriptor: | Fab ADD056 Heavy Chain, Fab ADD056 Light Chain, N-(4-{[(2,4-DIAMINOPTERIDIN-1-IUM-6-YL)METHYL](METHYL)AMINO}BENZOYL)-L-GLUTAMIC ACID | | Authors: | Longenecker, K.L, Judge, R.A, Gayda, S, Manoj, S, Saldana, S, Ruan, Q, Swift, K, Tetin, S. | | Deposit date: | 2014-01-09 | | Release date: | 2014-07-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Water channel in the binding site of a high affinity anti-methotrexate antibody.
Biochemistry, 53, 2014
|
|
4OCY
 
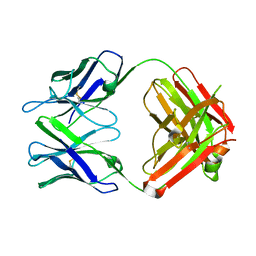 | | Fab for methotrexate (unbound apo) | | Descriptor: | Fab ADD058 Heavy Chain, Fab ADD058 Light Chain | | Authors: | Longenecker, K.L, Judge, R.A, Gayda, S, Manoj, S, Saldana, S, Ruan, Q, Swift, K, Tetin, S. | | Deposit date: | 2014-01-09 | | Release date: | 2014-07-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Water channel in the binding site of a high affinity anti-methotrexate antibody.
Biochemistry, 53, 2014
|
|
5DYO
 
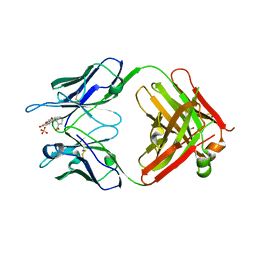 | | Fab43.1 complex with flourescein | | Descriptor: | 2-(6-HYDROXY-3-OXO-3H-XANTHEN-9-YL)-BENZOIC ACID, Fab 43.1 Heavy Chain, Fab 43.1 Light Chain, ... | | Authors: | Longenecker, K.L, Judge, R.A. | | Deposit date: | 2015-09-25 | | Release date: | 2016-01-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Three-dimensional structure, binding, and spectroscopic characteristics of the monoclonal antibody 43.1 directed to the carboxyphenyl moiety of fluorescein.
Biopolymers, 105, 2016
|
|
5EUE
 
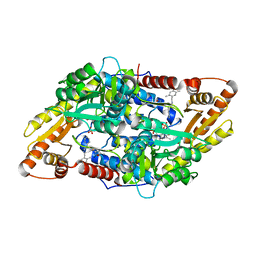 | | S1P Lyase Bacterial Surrogate bound to N-(2-((4-methoxy-2,5-dimethylbenzyl)amino)-1-phenylethyl)-5-methylisoxazole-3-carboxamide | | Descriptor: | PHOSPHATE ION, Putative sphingosine-1-phosphate lyase, ~{N}-[(1~{S})-2-[(4-methoxy-2,5-dimethyl-phenyl)methylamino]-1-phenyl-ethyl]-5-methyl-1,2-oxazole-3-carboxamide | | Authors: | Argiriadi, M.A, Banach, D, Radziejewska, E, Marchie, S, DiMauro, J, Dinges, J, Dominguez, E, Hutchins, C, Judge, R.A, Queeney, K, Wallace, G, Harris, C.M. | | Deposit date: | 2015-11-18 | | Release date: | 2016-03-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Creation of a S1P Lyase bacterial surrogate for structure-based drug design.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5EUD
 
 | | S1P Lyase Bacterial Surrogate bound to N-(1-(4-(3-hydroxyprop-1-yn-1-yl)phenyl)-2-((4-methoxy-2,5-dimethylbenzyl)amino)ethyl)-5-methylisoxazole-3-carboxamide | | Descriptor: | PHOSPHATE ION, Putative sphingosine-1-phosphate lyase, ~{N}-[(1~{S})-2-[(4-methoxy-2,5-dimethyl-phenyl)methylamino]-1-[4-(3-oxidanylprop-1-ynyl)phenyl]ethyl]-5-methyl-1,2-oxazole-3-carboxamide | | Authors: | Argiriadi, M.A, Banach, D, Radziejewska, E, Marchie, S, DiMauro, J, Dinges, J, Dominguez, E, Hutchins, C, Judge, R.A, Queeney, K, Wallace, G, Harris, C.M. | | Deposit date: | 2015-11-18 | | Release date: | 2016-03-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Creation of a S1P Lyase bacterial surrogate for structure-based drug design.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
7LAW
 
 | | crystal structure of GITR complex with GITR-L | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Tumor necrosis factor ligand superfamily member 18, Tumor necrosis factor receptor superfamily member 18 | | Authors: | Longenecker, K.L, Rogers, B, Bigelow, L, Judge, R.A, Alvarez, H. | | Deposit date: | 2021-01-07 | | Release date: | 2022-03-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.752 Å) | | Cite: | An anti-PD-1-GITR-L bispecific agonist induces GITR clustering-mediated T cell activation for cancer immunotherapy.
Nat Cancer, 3, 2022
|
|
2QF6
 
 | | HSP90 complexed with A56322 | | Descriptor: | 6-(3-BROMO-2-NAPHTHYL)-1,3,5-TRIAZINE-2,4-DIAMINE, Heat shock protein HSP 90-alpha | | Authors: | Park, C.H. | | Deposit date: | 2007-06-26 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Discovery and design of novel HSP90 inhibitors using multiple fragment-based design strategies.
Chem.Biol.Drug Des., 70, 2007
|
|
2QG2
 
 | | HSP90 complexed with A917985 | | Descriptor: | 3-({2-[(2-AMINO-6-METHYLPYRIMIDIN-4-YL)ETHYNYL]BENZYL}AMINO)-1,3-OXAZOL-2(3H)-ONE, Heat shock protein HSP 90-alpha | | Authors: | Park, C.H. | | Deposit date: | 2007-06-28 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery and design of novel HSP90 inhibitors using multiple fragment-based design strategies.
Chem.Biol.Drug Des., 70, 2007
|
|
2QG0
 
 | | HSP90 complexed with A943037 | | Descriptor: | Heat shock protein HSP 90-alpha, N-[(2-AMINO-6-METHYLPYRIMIDIN-4-YL)METHYL]-3-{[(E)-(2-OXODIHYDROFURAN-3(2H)-YLIDENE)METHYL]AMINO}BENZENESULFONAMIDE | | Authors: | Park, C.H. | | Deposit date: | 2007-06-28 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery and design of novel HSP90 inhibitors using multiple fragment-based design strategies.
Chem.Biol.Drug Des., 70, 2007
|
|
2QFO
 
 | | HSP90 complexed with A143571 and A516383 | | Descriptor: | (3E)-3-[(phenylamino)methylidene]dihydrofuran-2(3H)-one, 4-METHYL-6-(TRIFLUOROMETHYL)PYRIMIDIN-2-AMINE, Heat shock protein HSP 90-alpha | | Authors: | Park, C.H. | | Deposit date: | 2007-06-27 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Discovery and design of novel HSP90 inhibitors using multiple fragment-based design strategies.
CHEM.BIOL.DRUG DES., 70, 2007
|
|
4QVX
 
 | |
4LNC
 
 | | Neutron structure of the cyclic glucose bound Xylose Isomerase E186Q mutant | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, Xylose isomerase, ... | | Authors: | Munshi, P, Meilleur, F, Myles, D. | | Deposit date: | 2013-07-11 | | Release date: | 2014-02-12 | | Last modified: | 2024-02-28 | | Method: | NEUTRON DIFFRACTION (2.19 Å) | | Cite: | Neutron structure of the cyclic glucose-bound xylose isomerase E186Q mutant.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6PE7
 
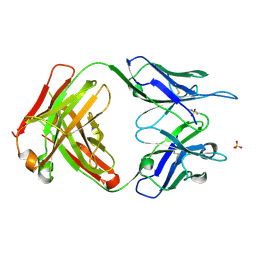 | | Crystal Structure of ABBV-323 FAB | | Descriptor: | FAB Heavy Chain, FAB Light chain, SULFATE ION | | Authors: | Argiriadi, M.A. | | Deposit date: | 2019-06-20 | | Release date: | 2019-08-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | CD40/anti-CD40 antibody complexes which illustrate agonist and antagonist structural switches.
BMC Mol Cell Biol, 20, 2019
|
|
6PE9
 
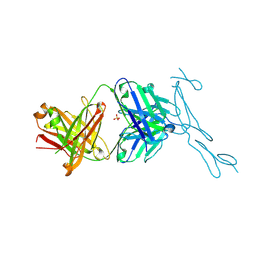 | | Crystal Structure of CD40 complexed to FAB516 | | Descriptor: | FAB Heavy chain, FAB Light chain, SULFATE ION, ... | | Authors: | Argiriadi, M.A. | | Deposit date: | 2019-06-20 | | Release date: | 2019-08-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | CD40/anti-CD40 antibody complexes which illustrate agonist and antagonist structural switches.
BMC Mol Cell Biol, 20, 2019
|
|
6PE8
 
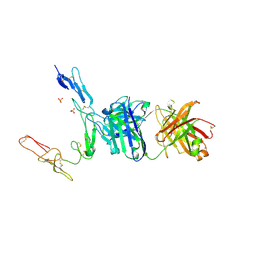 | | Crystal structure of CD40/ABBV-323 FAB complex | | Descriptor: | FAB Heavy chain, FAB Light chain, SULFATE ION, ... | | Authors: | Argiriadi, M.A. | | Deposit date: | 2019-06-20 | | Release date: | 2019-08-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | CD40/anti-CD40 antibody complexes which illustrate agonist and antagonist structural switches.
BMC Mol Cell Biol, 20, 2019
|
|
2EA2
 
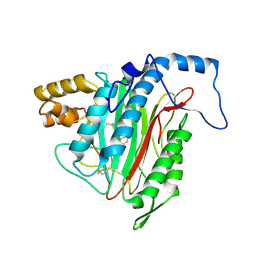 | | h-MetAP2 complexed with A773812 | | Descriptor: | 3-ETHYL-6-{[(4-FLUOROPHENYL)SULFONYL]AMINO}-2-METHYLBENZOIC ACID, MANGANESE (II) ION, Methionine aminopeptidase 2 | | Authors: | Park, C.H. | | Deposit date: | 2007-01-30 | | Release date: | 2008-02-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Lead optimization of methionine aminopeptidase-2 (MetAP2) inhibitors containing sulfonamides of 5,6-disubstituted anthranilic acids
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2EA4
 
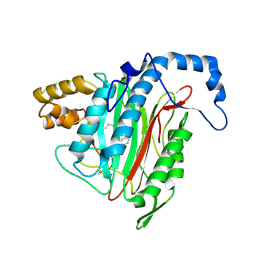 | | h-MetAP2 complexed with A797859 | | Descriptor: | 2-(2-AMINOETHOXY)-3-ETHYL-6-{[(4-FLUOROPHENYL)SULFONYL]AMINO}BENZOIC ACID, MANGANESE (II) ION, Methionine aminopeptidase 2 | | Authors: | Park, C.H. | | Deposit date: | 2007-01-30 | | Release date: | 2008-02-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Lead optimization of methionine aminopeptidase-2 (MetAP2) inhibitors containing sulfonamides of 5,6-disubstituted anthranilic acids
Bioorg.Med.Chem.Lett., 17, 2007
|
|
8DYG
 
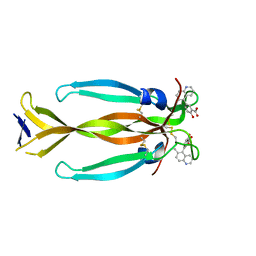 | | IL17A homodimer bound to Compound 7 | | Descriptor: | (5P)-2-hydroxy-5-(6-methylquinolin-5-yl)benzoic acid, Interleukin-17A | | Authors: | Argiriadi, M.A, Goedken, E.R. | | Deposit date: | 2022-08-04 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Identification and structure-based drug design of cell-active inhibitors of interleukin 17A at a novel C-terminal site.
Sci Rep, 12, 2022
|
|
8DYI
 
 | | IL17A homodimer bound to Compound 5 | | Descriptor: | (5P)-5-[5-(benzylamino)pyridin-3-yl]-N-[2-(morpholin-4-yl)ethyl]-1H-indazol-3-amine, Interleukin-17A | | Authors: | Argiriadi, M.A, Goedken, E.R. | | Deposit date: | 2022-08-04 | | Release date: | 2022-09-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Identification and structure-based drug design of cell-active inhibitors of interleukin 17A at a novel C-terminal site.
Sci Rep, 12, 2022
|
|
8DYH
 
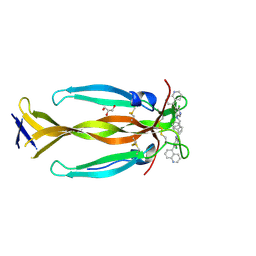 | | IL17A homodimer bound to Compound 6 | | Descriptor: | (5P)-N-benzyl-6-chloro-5-(quinolin-5-yl)pyridin-3-amine, GLYCEROL, Interleukin-17A | | Authors: | Argiriadi, M.A, Goedken, E.R. | | Deposit date: | 2022-08-04 | | Release date: | 2022-09-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Identification and structure-based drug design of cell-active inhibitors of interleukin 17A at a novel C-terminal site.
Sci Rep, 12, 2022
|
|
8DYF
 
 | | IL17A homodimer bound to Compound 10 | | Descriptor: | (5M)-3-[({2-[2-(2-{2-[2-({[(5M)-3-carboxy-5-(5,8-dihydroquinolin-4-yl)phenyl]amino}methyl)phenoxy]ethoxy}ethoxy)ethoxy]phenyl}methyl)amino]-5-(quinolin-4-yl)benzoic acid, Interleukin-17A | | Authors: | Argiriadi, M.A, Goedken, E.R. | | Deposit date: | 2022-08-04 | | Release date: | 2022-09-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Identification and structure-based drug design of cell-active inhibitors of interleukin 17A at a novel C-terminal site.
Sci Rep, 12, 2022
|
|
3OWB
 
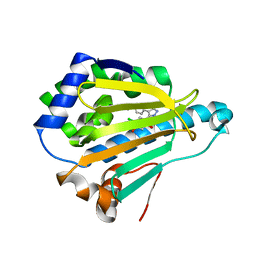 | | Crystal Structure of HSP90 with VER-49009 | | Descriptor: | 5-(5-CHLORO-2,4-DIHYDROXYPHENYL)-N-ETHYL-4-(4-METHOXYPHENYL)-1H-PYRAZOLE-3-CARBOXAMIDE, Heat shock protein HSP 90-alpha | | Authors: | Park, C.H. | | Deposit date: | 2010-09-17 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | N-aryl-benzimidazolones as novel small molecule HSP90 inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3OW6
 
 | | Crystal Structure of HSP90 with N-Aryl-benzimidazolone I | | Descriptor: | 1-(2,4-dihydroxyphenyl)-1,3-dihydro-2H-benzimidazol-2-one, Heat shock protein HSP 90-alpha | | Authors: | Park, C.H. | | Deposit date: | 2010-09-17 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | N-aryl-benzimidazolones as novel small molecule HSP90 inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3OWD
 
 | | Crystal Structure of HSP90 with N-Aryl-benzimidazolone II | | Descriptor: | Heat shock protein HSP 90-alpha, N-{[1-(5-chloro-2,4-dihydroxyphenyl)-2-oxo-2,3-dihydro-1H-benzimidazol-5-yl]methyl}naphthalene-1-sulfonamide | | Authors: | Park, C.H. | | Deposit date: | 2010-09-17 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | N-aryl-benzimidazolones as novel small molecule HSP90 inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2E9N
 
 | | Structure of h-CHK1 complexed with A767085 | | Descriptor: | 3-(4'-HYDROXYBIPHENYL-4-YL)-N-(4-HYDROXYCYCLOHEXYL)-1,4-DIHYDROINDENO[1,2-C]PYRAZOLE-6-CARBOXAMIDE, Serine/threonine-protein kinase Chk1 | | Authors: | Park, C. | | Deposit date: | 2007-01-26 | | Release date: | 2008-01-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of 1,4-dihydroindeno[1,2-c]pyrazoles as a novel class of potent and selective checkpoint kinase 1 inhibitors.
Bioorg.Med.Chem., 15, 2007
|
|
