2B4C
 
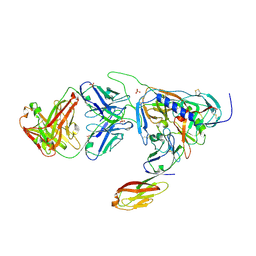 | | Crystal structure of HIV-1 JR-FL gp120 core protein containing the third variable region (V3) complexed with CD4 and the X5 antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, T-cell surface glycoprotein CD4, ... | | Authors: | Huang, C, Tang, M, Zhang, M.Y, Majeed, S, Montabana, E, Stanfield, R.L, Dimitrov, D.S, Korber, B, Sodroski, J, Wilson, I.A, Wyatt, R, Kwong, P.D. | | Deposit date: | 2005-09-23 | | Release date: | 2005-11-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of a V3-containing HIV-1 gp120 core.
Science, 310, 2005
|
|
3QC9
 
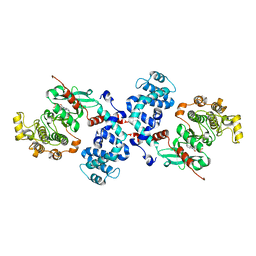 | |
5E5C
 
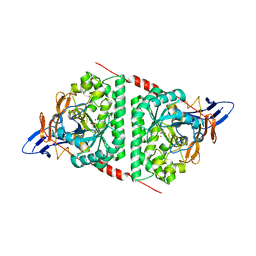 | | Crystal structure of dihydropyrimidinase from Pseudomonas aeruginosa PAO1 | | Descriptor: | D-hydantoinase/dihydropyrimidinase, ZINC ION | | Authors: | Huang, C.C, Huang, Y.H, Hsieh, Y.C, Tzeng, C.T, Chen, C.J, Huang, C.Y. | | Deposit date: | 2015-10-08 | | Release date: | 2016-09-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of dihydropyrimidinase from Pseudomonas aeruginosa PAO1: Insights into the molecular basis of formation of a dimer
Biochem.Biophys.Res.Commun., 478, 2016
|
|
5T8G
 
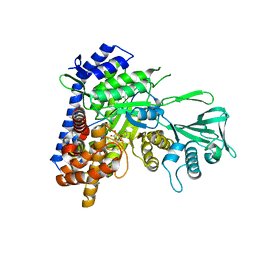 | |
6ZHY
 
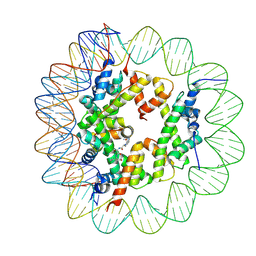 | | Cryo-EM structure of the regulatory linker of ALC1 bound to the nucleosome's acidic patch: hexasome class. | | Descriptor: | Chromodomain-helicase-DNA-binding protein 1-like, DNA (110-MER) Widom 601 sequence, Histone H2A type 1, ... | | Authors: | Bacic, L, Gaullier, G, Deindl, S. | | Deposit date: | 2020-06-24 | | Release date: | 2020-12-23 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanistic Insights into Regulation of the ALC1 Remodeler by the Nucleosome Acidic Patch.
Cell Rep, 33, 2020
|
|
4R4B
 
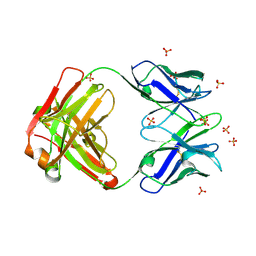 | | Crystal structure of the anti-hiv-1 antibody 2.2c | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FAB 2.2C HEAVY CHAIN, FAB 2.2C LIGHT CHAIN, ... | | Authors: | McLellan, J.S, Acharya, P, Huang, C.-C, Robinson, J, Kwong, P.D. | | Deposit date: | 2014-08-19 | | Release date: | 2014-09-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Structural Definition of an Antibody-Dependent Cellular Cytotoxicity Response Implicated in Reduced Risk for HIV-1 Infection.
J.Virol., 88, 2014
|
|
4R4H
 
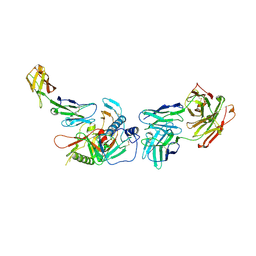 | | Crystal structure of non-neutralizing, A32-like antibody 2.2c in complex with HIV-1 Env gp120 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody 2.2c, Heavy chain, ... | | Authors: | Mclellan, J, Acharya, P, Huang, C.-C, Kwong, P.D. | | Deposit date: | 2014-08-19 | | Release date: | 2014-09-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (4.28 Å) | | Cite: | Structural Definition of an Antibody-Dependent Cellular Cytotoxicity Response Implicated in Reduced Risk for HIV-1 Infection.
J.Virol., 88, 2014
|
|
4R4F
 
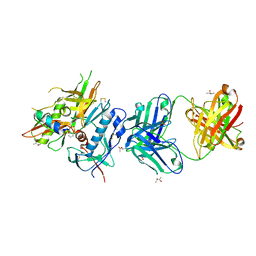 | | Crystal structure of non-neutralizing, A32-like antibody 2.2c in complex with HIV-1 YU2 gp120 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody 2.2c LIGHT CHAIN, ... | | Authors: | Acharya, P, Kwong, P.D. | | Deposit date: | 2014-08-19 | | Release date: | 2014-09-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.514 Å) | | Cite: | Structural Definition of an Antibody-Dependent Cellular Cytotoxicity Response Implicated in Reduced Risk for HIV-1 Infection.
J.Virol., 88, 2014
|
|
4R4N
 
 | | Crystal structure of the anti-hiv-1 antibody 2.2c in complex with hiv-1 93ug037 gp120 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody 2.2c LIGHT CHAIN, Antibody 2.2c heavy CHAIN, ... | | Authors: | Acharya, P, Louder, R, Kwong, P.D. | | Deposit date: | 2014-08-19 | | Release date: | 2014-11-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.56 Å) | | Cite: | Structural Definition of an Antibody-Dependent Cellular Cytotoxicity Response Implicated in Reduced Risk for HIV-1 Infection.
J.Virol., 88, 2014
|
|
4H8W
 
 | | Crystal structure of non-neutralizing and ADCC-potent antibody N5-i5 in complex with HIV-1 clade A/E gp120 and sCD4. | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, FAB HEAVY CHAIN OF ADCC AND NON-NEUTRALIZING ANTI-HIV-1 ANTIBODY N5-I5, ... | | Authors: | Tolbert, W.D, Acharya, P, Kwong, P.D, Pazgier, M. | | Deposit date: | 2012-09-24 | | Release date: | 2014-04-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Definition of an Antibody-Dependent Cellular Cytotoxicity Response Implicated in Reduced Risk for HIV-1 Infection.
J.Virol., 88, 2014
|
|
8HFD
 
 | | Crystal structure of allantoinase from E. coli BL21 | | Descriptor: | Allantoinase, DI(HYDROXYETHYL)ETHER, ZINC ION | | Authors: | Lin, E.S, Huang, H.Y, Yang, P.C, Liu, H.W, Huang, C.Y. | | Deposit date: | 2022-11-10 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal Structure of Allantoinase from Escherichia coli BL21: A Molecular Insight into a Role of the Active Site Loops in Catalysis.
Molecules, 28, 2023
|
|
8C21
 
 | |
8C1Z
 
 | |
8C1W
 
 | |
8C20
 
 | |
9FCZ
 
 | | Coxsackievirus A9 bound with compound 17 (CL301) | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Plavec, Z, Butcher, S.J, Mitchell, C, Buckner, C. | | Deposit date: | 2024-05-16 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | SAR Analysis of Novel Coxsackie virus A9 Capsid Binders.
J.Med.Chem., 67, 2024
|
|
9EVD
 
 | | In situ structure of the peripheral stalk of the mitochondrial ATPsynthase in whole Polytomella cells | | Descriptor: | ASA-10: Polytomella F-ATP synthase associated subunit 10, ASA-9: Polytomella F-ATP synthase associated subunit 9, ATP synthase associated protein ASA1, ... | | Authors: | Dietrich, L, Agip, A.N.A, Kuehlbrandt, W. | | Deposit date: | 2024-03-29 | | Release date: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | In situ structure and rotary states of mitochondrial ATP synthase in whole Polytomella cells.
Science, 385, 2024
|
|
9EXI
 
 | | Coxsackievirus A9 bound with compound 14 (CL275) | | Descriptor: | 4-[(4-methylpiperazin-1-yl)methyl]-N-[[4-(trifluoromethyl)phenyl]methyl]aniline, Capsid protein VP1, Capsid protein VP2, ... | | Authors: | Plavec, Z, Butcher, S.J, Mitchell, C, Buckner, C. | | Deposit date: | 2024-04-08 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.31 Å) | | Cite: | SAR Analysis of Novel Coxsackie virus A9 Capsid Binders.
J.Med.Chem., 67, 2024
|
|
9FP5
 
 | | Coxsackievirus A9 bound with CL213. | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Plavec, Z, Butcher, S.J, Mitchell, C, Buckner, C. | | Deposit date: | 2024-06-13 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | SAR Analysis of Novel Coxsackie virus A9 Capsid Binders.
J.Med.Chem., 67, 2024
|
|
9FA9
 
 | | Coxsackievirus A9 bound with compound 16 (CL298) | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Plavec, Z, Butcher, S.J, Mitchell, C, Buckner, C. | | Deposit date: | 2024-05-10 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | SAR Analysis of Novel Coxsackie virus A9 Capsid Binders.
J.Med.Chem., 67, 2024
|
|
9FGN
 
 | | Coxsackievirus A9 bound with compound 18 (CL304) | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Plavec, Z, Butcher, S.J, Mitchell, C, Buckner, C. | | Deposit date: | 2024-05-24 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | SAR Analysis of Novel Coxsackie virus A9 Capsid Binders.
J.Med.Chem., 67, 2024
|
|
2I60
 
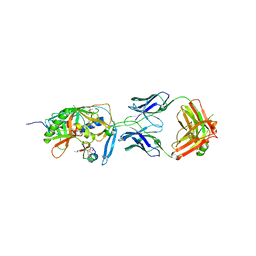 | | Crystal structure of [Phe23]M47, a scorpion-toxin mimic of CD4, in complex with HIV-1 YU2 GP120 envelope glycoprotein and anti-HIV-1 antibody 17B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ANTIBODY 17B HEAVY CHAIN, ANTIBODY 17B LIGHT CHAIN, ... | | Authors: | Huang, C.-C, Kwong, P.D. | | Deposit date: | 2006-08-26 | | Release date: | 2006-10-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Combinatorial optimization of a CD4-mimetic miniprotein and cocrystal structures with HIV-1 gp120 envelope glycoprotein.
J.Mol.Biol., 382, 2008
|
|
2I5Y
 
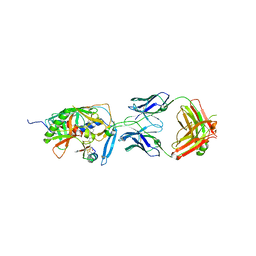 | | Crystal structure of CD4M47, a scorpion-toxin mimic of CD4, in complex with HIV-1 YU2 GP120 envelope glycoprotein and anti-HIV-1 antibody 17B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody 17B Heavy chain, Antibody 17B Light chain, ... | | Authors: | Huang, C.-C, Kwong, P.D. | | Deposit date: | 2006-08-26 | | Release date: | 2006-10-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Combinatorial optimization of a CD4-mimetic miniprotein and cocrystal structures with HIV-1 gp120 envelope glycoprotein.
J.Mol.Biol., 382, 2008
|
|
8RJB
 
 | |
8S7J
 
 | | Coxsackievirus A9 bound with compound 20 (CL300) | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Plavec, Z, Butcher, S.J, Mitchell, C, Buckner, C. | | Deposit date: | 2024-03-01 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.26 Å) | | Cite: | SAR Analysis of Novel Coxsackie virus A9 Capsid Binders.
J.Med.Chem., 67, 2024
|
|
