8TTL
 
 | | AT8-Phosphomimetic Tau Filaments (Full-length, Cofactor-Free 0N4R Tau S202E, T205E, S208E) | | Descriptor: | Microtubule-associated protein tau | | Authors: | El Mammeri, N, Dregni, A.J, Duan, P, Hong, M. | | Deposit date: | 2023-08-14 | | Release date: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structures of AT8 and PHF1 phosphomimetic tau: Insights into the posttranslational modification code of tau aggregation.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6PVR
 
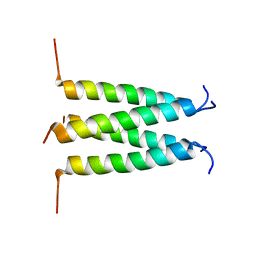 | | Influenza B M2 Proton Channel in the Closed State - SSNMR Structure at pH 7.5 | | Descriptor: | BM2 protein | | Authors: | Mandala, V.S, Loftis, A.R, Shcherbakov, A.S, Pentelute, B.L, Hong, M. | | Deposit date: | 2019-07-21 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Atomic structures of closed and open influenza B M2 proton channel reveal the conduction mechanism.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7JK8
 
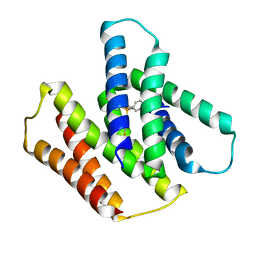 | | EmrE S64V mutant bound to tetra(4-fluorophenyl)phosphonium at pH 5.8 | | Descriptor: | Multidrug SMR transporter, tetrakis(4-fluorophenyl)phosphanium | | Authors: | Shcherbakov, A.A, Hisao, G, Mandala, V.S, Thomas, N.E, Soltani, M, Salter, E.A, Davis Jr, J.H, Henzler-Wildman, K.A, Hong, M. | | Deposit date: | 2020-07-27 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Structure and dynamics of the drug-bound bacterial transporter EmrE in lipid bilayers.
Nat Commun, 12, 2021
|
|
5IE9
 
 | |
2KQT
 
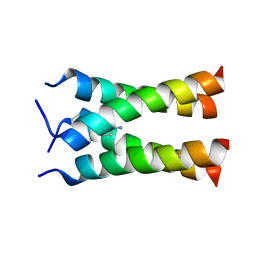 | | Solid-state NMR structure of the M2 transmembrane peptide of the influenza A virus in DMPC lipid bilayers bound to deuterated amantadine | | Descriptor: | (3S,5S,7S)-tricyclo[3.3.1.1~3,7~]decan-1-amine, M2 protein | | Authors: | Cady, S.D, Schmidt-Rohr, K, Wang, J, Soto, C.S, DeGrado, W.F, Hong, M. | | Deposit date: | 2009-11-18 | | Release date: | 2010-02-09 | | Last modified: | 2024-05-08 | | Method: | SOLID-STATE NMR | | Cite: | Structure of the amantadine binding site of influenza M2 proton channels in lipid bilayers
Nature, 463, 2010
|
|
5UGK
 
 | | Zinc-Binding Structure of a Catalytic Amyloid from Solid-State NMR Spectroscopy | | Descriptor: | ILE-HIS-VAL-HIS-LEU-GLN-ILE, ZINC ION | | Authors: | Lee, M, Wang, T, Makhlynets, O.V, Wu, Y, Polizzi, N, Wu, H, Gosavi, P.M, Korendovych, I.V, DeGrado, W.F, Hong, M. | | Deposit date: | 2017-01-09 | | Release date: | 2017-05-31 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Zinc-binding structure of a catalytic amyloid from solid-state NMR.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5H20
 
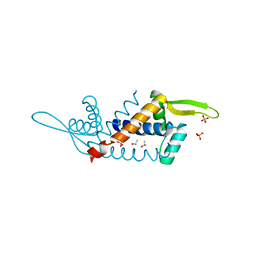 | | X-ray structure of PadR-like Transcription factor from bacteroid fragilis | | Descriptor: | ISOPROPYL ALCOHOL, PHOSPHATE ION, Putative PadR-family transcriptional regulatory protein, ... | | Authors: | Lee, C, Hong, M. | | Deposit date: | 2016-10-13 | | Release date: | 2017-03-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and functional analysis of BF2549, a PadR-like transcription factor from Bacteroides fragilis.
Biochem. Biophys. Res. Commun., 483, 2017
|
|
6DLN
 
 | |
6NZN
 
 | | Dimer-of-dimer amyloid fibril structure of glucagon | | Descriptor: | Glucagon | | Authors: | Gelenter, M.D, Smith, K.J, Liao, S.Y, Mandala, V.S, Dregni, A.J, Lamm, M.S, Tian, Y, Wei, X, Pochan, D.J, Tucker, T.J, Su, Y, Hong, M. | | Deposit date: | 2019-02-14 | | Release date: | 2019-06-05 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | The peptide hormone glucagon forms amyloid fibrils with two coexisting beta-strand conformations.
Nat.Struct.Mol.Biol., 26, 2019
|
|
3MTX
 
 | | Crystal structure of chicken MD-1 | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, GLYCEROL, Protein MD-1, ... | | Authors: | Yoon, S.I, Hong, M, Han, G.W, Wilson, I.A. | | Deposit date: | 2010-05-01 | | Release date: | 2010-06-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of soluble MD-1 and its interaction with lipid IVa.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3MU3
 
 | | Crystal structure of chicken MD-1 complexed with lipid IVa | | Descriptor: | (R)-((2R,3S,4R,5R,6R)-3-HYDROXY-2-(HYDROXYMETHYL)-5-((R)-3-HYDROXYTETRADECANAMIDO)-6-(PHOSPHONOOXY)TETRAHYDRO-2H-PYRAN-4-YL) 3-HYDROXYTETRADECANOATE, 2-deoxy-3-O-[(3R)-3-hydroxytetradecanoyl]-2-{[(3R)-3-hydroxytetradecanoyl]amino}-4-O-phosphono-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Yoon, S.I, Hong, M, Han, G.W, Wilson, I.A. | | Deposit date: | 2010-05-01 | | Release date: | 2010-06-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of soluble MD-1 and its interaction with lipid IVa.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3RG1
 
 | | Crystal structure of the RP105/MD-1 complex | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, CD180 molecule, LY86 protein, ... | | Authors: | Yoon, S.I, Hong, M, Wilson, I.A. | | Deposit date: | 2011-04-07 | | Release date: | 2011-08-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | An unusual dimeric structure and assembly for TLR4 regulator RP105-MD-1.
Nat.Struct.Mol.Biol., 18, 2011
|
|
2HVC
 
 | | The Crystal Structure of Ligand-binding Domain (LBD) of human Androgen Receptor in Complex with a selective modulator LGD2226 | | Descriptor: | 6-[BIS(2,2,2-TRIFLUOROETHYL)AMINO]-4-(TRIFLUOROMETHYL)QUINOLIN-2(1H)-ONE, Androgen receptor | | Authors: | Wang, F, Liu, X.-Q, Li, H, Liang, K.-N, Miner, J.N, Hong, M, Kallel, E.A, van Oeveren, A, Zhi, L, Jiang, T. | | Deposit date: | 2006-07-28 | | Release date: | 2007-07-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the ligand-binding domain (LBD) of human androgen receptor in complex with a selective modulator LGD2226
ACTA CRYSTALLOGR.,SECT.F, 62, 2006
|
|
5X12
 
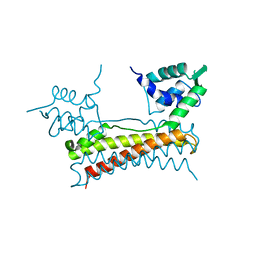 | | Crystal structure of Bacillus subtilis PadR | | Descriptor: | Transcriptional regulator | | Authors: | Park, S.C, Kwak, Y.M, Song, W.S, Hong, M, Yoon, S.I. | | Deposit date: | 2017-01-24 | | Release date: | 2017-11-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of effector and operator recognition by the phenolic acid-responsive transcriptional regulator PadR
Nucleic Acids Res., 45, 2017
|
|
5X11
 
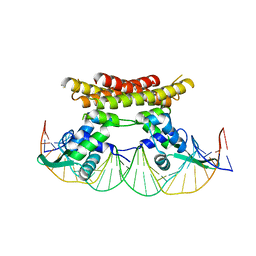 | | Crystal structure of Bacillus subtilis PadR in complex with operator DNA | | Descriptor: | DNA (28-MER), Transcriptional regulator | | Authors: | Park, S.C, Kwak, Y.M, Song, W.S, Hong, M, Yoon, S.I. | | Deposit date: | 2017-01-24 | | Release date: | 2017-11-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis of effector and operator recognition by the phenolic acid-responsive transcriptional regulator PadR
Nucleic Acids Res., 45, 2017
|
|
5Z7Q
 
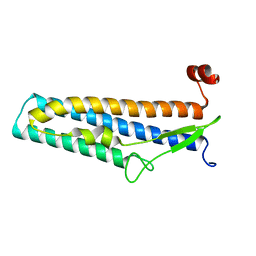 | | Crystal structure of Bacillus cereus flagellin | | Descriptor: | Flagellin | | Authors: | Kim, M, Hong, M. | | Deposit date: | 2018-01-30 | | Release date: | 2018-05-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of Bacillus cereus flagellin and structure-guided fusion-protein designs
Sci Rep, 8, 2018
|
|
5X14
 
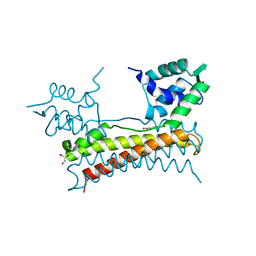 | | Crystal structure of Bacillus subtilis PadR in complex with ferulic acid | | Descriptor: | 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, GLYCEROL, Transcriptional regulator | | Authors: | Park, S.C, Kwak, Y.M, Song, W.S, Hong, M, Yoon, S.I. | | Deposit date: | 2017-01-24 | | Release date: | 2017-11-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural basis of effector and operator recognition by the phenolic acid-responsive transcriptional regulator PadR
Nucleic Acids Res., 45, 2017
|
|
5X13
 
 | | Crystal structure of Bacillus subtilis PadR in complex with p-coumaric acid | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, GLYCEROL, Transcriptional regulator | | Authors: | Park, S.C, Kwak, Y.M, Song, W.S, Hong, M, Yoon, S.I. | | Deposit date: | 2017-01-24 | | Release date: | 2017-11-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of effector and operator recognition by the phenolic acid-responsive transcriptional regulator PadR
Nucleic Acids Res., 45, 2017
|
|
2MUZ
 
 | | ssNMR structure of a designed rocker protein | | Descriptor: | designed rocker protein | | Authors: | Wang, T, Joh, N, Wu, Y, DeGrado, W.F, Hong, M. | | Deposit date: | 2014-09-18 | | Release date: | 2014-12-24 | | Last modified: | 2015-01-14 | | Method: | SOLUTION NMR | | Cite: | De novo design of a transmembrane Zn2+-transporting four-helix bundle.
Science, 346, 2014
|
|
2KHT
 
 | | NMR Structure of human alpha defensin HNP-1 | | Descriptor: | Neutrophil defensin 1 | | Authors: | Zhang, Y, Li, S, Doherty, T.F, Lubkowski, J, Lu, W, Li, J, Barinka, C, Hong, M. | | Deposit date: | 2009-04-11 | | Release date: | 2010-02-09 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Resonance assignment and three-dimensional structure determination of a human alpha-defensin, HNP-1, by solid-state NMR.
J.Mol.Biol., 397, 2010
|
|
5Y8T
 
 | | Crystal structure of Bacillus subtilis PadR in complex with p-coumaric acid | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Transcriptional regulator | | Authors: | Park, S.C, Kwak, Y.M, Song, W.S, Hong, M, Yoon, S.I. | | Deposit date: | 2017-08-21 | | Release date: | 2017-11-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of effector and operator recognition by the phenolic acid-responsive transcriptional regulator PadR.
Nucleic Acids Res., 45, 2017
|
|
6ABT
 
 | | Crystal structure of transcription factor from Listeria monocytogenes | | Descriptor: | PadR family transcriptional regulator | | Authors: | Lee, C, Hong, M. | | Deposit date: | 2018-07-23 | | Release date: | 2019-06-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based molecular characterization and regulatory mechanism of the LftR transcription factor from Listeria monocytogenes: Conformational flexibilities and a ligand-induced regulatory mechanism.
Plos One, 14, 2019
|
|
6ABQ
 
 | | Crystal structure of transcription factor from Listeria monocytogenes | | Descriptor: | CHLORIDE ION, PadR family transcriptional regulator | | Authors: | Lee, C, Hong, M. | | Deposit date: | 2018-07-23 | | Release date: | 2019-06-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based molecular characterization and regulatory mechanism of the LftR transcription factor from Listeria monocytogenes: Conformational flexibilities and a ligand-induced regulatory mechanism.
Plos One, 14, 2019
|
|
5WYR
 
 | | Crystal structure and catalytic mechanism of the essential m1G37 tRNA methyltransferase TrmD from Pseudomonas aeruginosa | | Descriptor: | SINEFUNGIN, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Jaroensuk, J, Liew, C.W, Atichartpongkul, S, Chionh, Y.H, Wong, Y.H, Zhong, W.H, McBee, M.E, Thongdee, N, Prestwich, E.G, DeMott, M.S, Mongkolsuk, S, Dedon, P.C, Lescar, J, Fuangthong, M. | | Deposit date: | 2017-01-15 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure and catalytic mechanism of the essential m1G37 tRNA methyltransferase TrmD fromPseudomonas aeruginosa.
Rna, 2019
|
|
5WYQ
 
 | | Crystal Structure and catalytic mechanism of the essential m1G37 tRNA methyltransferase TrmD from Pseudomonas aeruginosa | | Descriptor: | S-ADENOSYLMETHIONINE, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Jaroensuk, J, Liew, C.W, Atichartpongkul, S, Chionh, Y.H, Wong, Y.H, Zhong, W, McBee, M.E, Maenpuen, S, Prestwich, E.G, DeMott, M.S, Mongkolsuk, S, Dedon, P.C, Lescar, J, Fuangthong, M, Chaiyen, P. | | Deposit date: | 2017-01-15 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure and catalytic mechanism of the essential m1G37 tRNA methyltransferase TrmD fromPseudomonas aeruginosa.
Rna, 2019
|
|
