3S27
 
 | |
3S29
 
 | |
3PQE
 
 | |
3S28
 
 | | The crystal structure of sucrose synthase-1 in complex with a breakdown product of the UDP-glucose | | Descriptor: | 1,5-anhydro-D-arabino-hex-1-enitol, 1,5-anhydro-D-fructose, MALONIC ACID, ... | | Authors: | Zheng, Y, Garavito, R.M. | | Deposit date: | 2011-05-16 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Structure of Sucrose Synthase-1 from Arabidopsis thaliana and Its Functional Implications.
J.Biol.Chem., 286, 2011
|
|
3PQD
 
 | |
1I2C
 
 | | CRYSTAL STRUCTURE OF MUTANT T145A SQD1 PROTEIN COMPLEX WITH NAD AND UDP-GLUCOSE | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, SULFOLIPID BIOSYNTHESIS PROTEIN SQD1, ... | | Authors: | Theisen, M.J, Sanda, S.L, Ginell, S.L, Benning, C, Garavito, R.M. | | Deposit date: | 2001-02-07 | | Release date: | 2003-07-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Characterization of the Active Site of UDP-sulfoquinovose Synthase: Formation of the Sulfonic Acid Product in the Crystalline State.
To be Published
|
|
1IIR
 
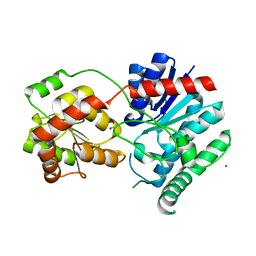 | | Crystal Structure of UDP-glucosyltransferase GtfB | | Descriptor: | MAGNESIUM ION, SULFATE ION, glycosyltransferase GtfB | | Authors: | Mulichak, A.M, Losey, H.C, Walsh, C.T, Garavito, R.M. | | Deposit date: | 2001-04-24 | | Release date: | 2001-07-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the UDP-glucosyltransferase GtfB that modifies the heptapeptide aglycone in the biosynthesis of vancomycin group antibiotics.
Structure, 9, 2001
|
|
1I24
 
 | | HIGH RESOLUTION CRYSTAL STRUCTURE OF THE WILD-TYPE PROTEIN SQD1, WITH NAD AND UDP-GLUCOSE | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, SULFOLIPID BIOSYNTHESIS PROTEIN SQD1, ... | | Authors: | Theisen, M.J, Sanda, S.L, Ginell, S.L, Benning, C, Garavito, R.M. | | Deposit date: | 2001-02-05 | | Release date: | 2003-07-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Characterization of the Active Site of Udp-Sulfoquinovose Synthase: Formation of the Sulfonic Acid Product in the Crystalline State
To be Published
|
|
1I2B
 
 | | CRYSTAL STRUCTURE OF MUTANT T145A SQD1 PROTEIN COMPLEX WITH NAD AND UDP-SULFOQUINOVOSE/UDP-GLUCOSE | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, SULFOLIPID BIOSYNTHESIS PROTEIN SQD1, ... | | Authors: | Theisen, M.J, Sanda, S.L, Ginell, S.L, Benning, C, Garavito, R.M. | | Deposit date: | 2001-02-07 | | Release date: | 2003-07-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Characterization of the Active Site of UDP-sulfoquinovose Synthase: Formation of the Sulfonic Acid Product in the Crystalline State
To be Published
|
|
1DIY
 
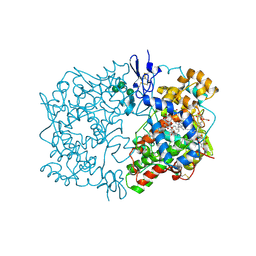 | | CRYSTAL STRUCTURE OF ARACHIDONIC ACID BOUND IN THE CYCLOOXYGENASE ACTIVE SITE OF PGHS-1 | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ARACHIDONIC ACID, ... | | Authors: | Malkowski, M.G, Ginell, S.L, Smith, W.L, Garavito, R.M. | | Deposit date: | 1999-11-30 | | Release date: | 2000-09-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The productive conformation of arachidonic acid bound to prostaglandin synthase.
Science, 289, 2000
|
|
1BG3
 
 | |
1BDG
 
 | |
1PNV
 
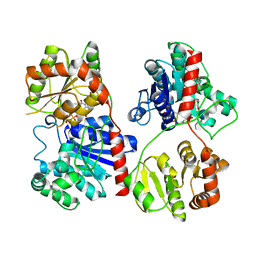 | | Crystal Structure of TDP-epi-Vancosaminyltransferase GtfA in complexes with TDP and Vancomycin | | Descriptor: | GLYCOSYLTRANSFERASE GTFA, THYMIDINE-5'-DIPHOSPHATE, VANCOMYCIN, ... | | Authors: | Mulichak, A.M, Losey, H.C, Lu, W, Wawrzak, Z, Walsh, C.T, Garavito, R.M. | | Deposit date: | 2003-06-13 | | Release date: | 2003-08-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Tdp-Epi-Vancosaminyltransferase Gtfa from the Chloroeremomycin Biosynthetic Pathway.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1PN3
 
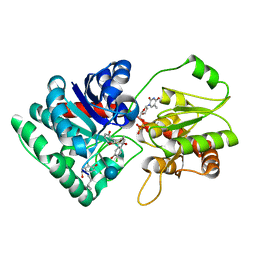 | | Crystal Structure of TDP-epi-Vancosaminyltransferase GtfA in complexes with TDP and the acceptor substrate DVV. | | Descriptor: | DESVANCOSAMINYL VANCOMYCIN, GLYCOSYLTRANSFERASE GTFA, THYMIDINE-5'-DIPHOSPHATE, ... | | Authors: | Mulichak, A.M, Losey, H.C, Lu, W, Wawrzak, Z, Walsh, C.T, Garavito, R.M. | | Deposit date: | 2003-06-12 | | Release date: | 2003-08-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Tdp-Epi-Vancosaminyltransferase Gtfa from the Chloroeremomycin Biosynthetic Pathway.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1N7H
 
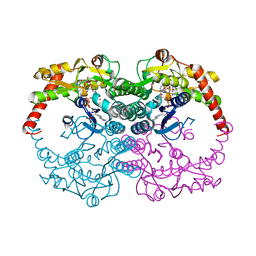 | | Crystal Structure of GDP-mannose 4,6-dehydratase ternary complex with NADPH and GDP | | Descriptor: | GDP-D-mannose-4,6-dehydratase, GUANOSINE-5'-DIPHOSPHATE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Mulichak, A.M, Bonin, C.P, Reiter, W.-D, Garavito, R.M. | | Deposit date: | 2002-11-14 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of the MUR1 GDP-mannose
4,6-dehydratase from A. thaliana:
Implications for ligand binding and specificity.
Biochemistry, 41, 2002
|
|
1N7G
 
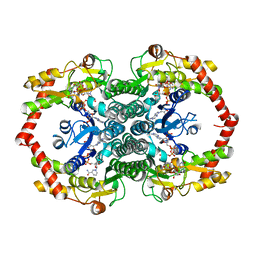 | | Crystal Structure of the GDP-mannose 4,6-dehydratase ternary complex with NADPH and GDP-rhamnose. | | Descriptor: | GDP-D-mannose-4,6-dehydratase, GUANOSINE-5'-DIPHOSPHATE-RHAMNOSE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Mulichak, A.M, Bonin, C.P, Reiter, W.-D, Garavito, R.M. | | Deposit date: | 2002-11-14 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of the MUR1 GDP-mannose 4,6-dehydratase
from A. thaliana: Implications for ligand binding and
specificity.
Biochemistry, 41, 2002
|
|
1QRR
 
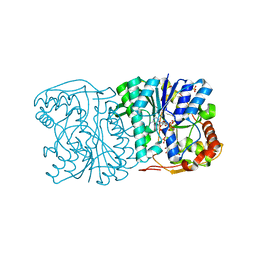 | | CRYSTAL STRUCTURE OF SQD1 PROTEIN COMPLEX WITH NAD AND UDP-GLUCOSE | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, URIDINE-5'-DIPHOSPHATE-GLUCOSE, ... | | Authors: | Mulichak, A.M, Theisen, M.J, Essigmann, B, Benning, C, Garavito, R.M. | | Deposit date: | 1999-06-15 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of SQD1, an enzyme involved in the biosynthesis of the plant sulfolipid headgroup donor UDP-sulfoquinovose.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1RRV
 
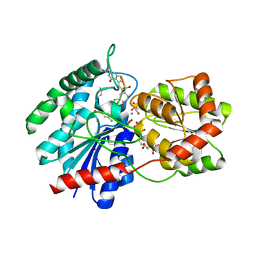 | | X-ray crystal structure of TDP-vancosaminyltransferase GtfD as a complex with TDP and the natural substrate, desvancosaminyl vancomycin. | | Descriptor: | DESVANCOSAMINYL VANCOMYCIN, GLYCEROL, GLYCOSYLTRANSFERASE GTFD, ... | | Authors: | Mulichak, A.M, Lu, W, Losey, H.C, Walsh, C.T, Garavito, R.M. | | Deposit date: | 2003-12-09 | | Release date: | 2004-05-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Vancosaminyltransferase Gtfd from the Vancomycin Biosynthetic Pathway: Interactions with Acceptor and Nucleotide Ligands
Biochemistry, 43, 2004
|
|
4GPD
 
 | |
1OPF
 
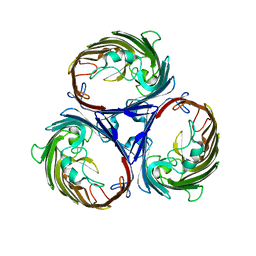 | |
1GPD
 
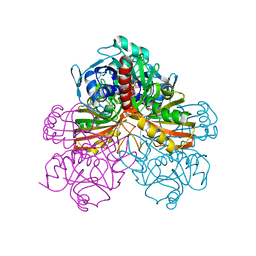 | | STUDIES OF ASYMMETRY IN THE THREE-DIMENSIONAL STRUCTURE OF LOBSTER D-GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE | | Descriptor: | D-GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Moras, D, Olsen, K.W, Sabesan, M.N, Buehner, M, Ford, G.C, Rossmann, M.G. | | Deposit date: | 1975-07-01 | | Release date: | 1977-02-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Studies of asymmetry in the three-dimensional structure of lobster D-glyceraldehyde-3-phosphate dehydrogenase.
J.Biol.Chem., 250, 1975
|
|
8E7Y
 
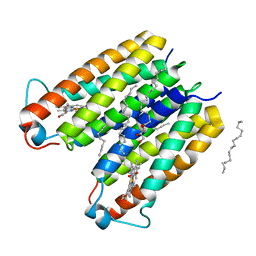 | | RsTSPO A138F with two heme bound | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, PROTOPORPHYRIN IX CONTAINING FE, Tryptophan-rich sensory protein | | Authors: | Liu, J, Hiser, C, Li, F, Garavito, R, Ferguson-Miller, S. | | Deposit date: | 2022-08-25 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | New TSPO Crystal Structures of Mutant and Heme-Bound Forms with Altered Flexibility, Ligand Binding, and Porphyrin Degradation Activity.
Biochemistry, 62, 2023
|
|
8E7Z
 
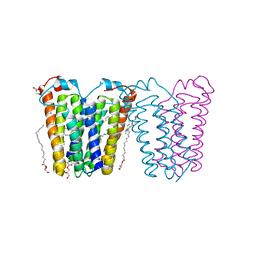 | | RsTSPO mutant -A138F | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, DI(HYDROXYETHYL)ETHER, Tryptophan-rich sensory protein | | Authors: | Liu, J, Hiser, C, Li, F, Garavito, R, Ferguson-Miller, S. | | Deposit date: | 2022-08-25 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | New TSPO Crystal Structures of Mutant and Heme-Bound Forms with Altered Flexibility, Ligand Binding, and Porphyrin Degradation Activity.
Biochemistry, 62, 2023
|
|
8E7W
 
 | | RsTSPO A139T with Heme | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-2-[(6E)-HEXADEC-6-ENOYLOXY]PROPYL (8E)-OCTADEC-8-ENOATE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Liu, J, Hiser, C, Li, F, Garavito, R, Ferguson-Miller, S. | | Deposit date: | 2022-08-25 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | New TSPO Crystal Structures of Mutant and Heme-Bound Forms with Altered Flexibility, Ligand Binding, and Porphyrin Degradation Activity.
Biochemistry, 62, 2023
|
|
8E7X
 
 | | RsTSPO A138F with one Heme bound | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, PROTOPORPHYRIN IX CONTAINING FE, Tryptophan-rich sensory protein | | Authors: | Liu, J, Hiser, C, Li, F, Garavito, R, Ferguson-Miller, S. | | Deposit date: | 2022-08-25 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | New TSPO Crystal Structures of Mutant and Heme-Bound Forms with Altered Flexibility, Ligand Binding, and Porphyrin Degradation Activity.
Biochemistry, 62, 2023
|
|
