7WPV
 
 | | Fab14 - a SARS-CoV2 RBD neutralising antibody | | Descriptor: | Fab14 heavy chain, Fab14 light chain | | Authors: | Lin, J.Q, El Sahili, A, Lescar, J. | | Deposit date: | 2022-01-24 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Engineering SARS-CoV-2 specific cocktail antibodies into a bispecific format improves neutralizing potency and breadth.
Nat Commun, 13, 2022
|
|
7WPH
 
 | | SARS-CoV2 RBD bound to Fab06 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FAB06 light chain, Fab06 heavy chain, ... | | Authors: | Lin, J.Q, El Sahili, A, Lescar, J. | | Deposit date: | 2022-01-23 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Engineering SARS-CoV-2 specific cocktail antibodies into a bispecific format improves neutralizing potency and breadth.
Nat Commun, 13, 2022
|
|
5YVW
 
 | |
5YVU
 
 | |
7DIJ
 
 | | Falcilysin in complex with MK-4815 | | Descriptor: | 1,2-ETHANEDIOL, 2-(aminomethyl)-3,5-ditert-butyl-phenol, ACETATE ION, ... | | Authors: | Lin, J.Q, El Sahili, A, Lescar, J. | | Deposit date: | 2020-11-19 | | Release date: | 2021-12-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of an inhibitory pocket in falcilysin provides a new avenue for malaria drug development.
Cell Chem Biol, 31, 2024
|
|
7DI7
 
 | | Falcilysin in complex with chloroquine | | Descriptor: | ACETATE ION, Falcilysin, N4-(7-CHLORO-QUINOLIN-4-YL)-N1,N1-DIETHYL-PENTANE-1,4-DIAMINE, ... | | Authors: | Lin, J.Q, El Sahili, A, Lescar, J. | | Deposit date: | 2020-11-18 | | Release date: | 2021-12-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Identification of an inhibitory pocket in falcilysin provides a new avenue for malaria drug development.
Cell Chem Biol, 31, 2024
|
|
7DIA
 
 | | Falcilysin in complex with mefloquine | | Descriptor: | (S)-[2,8-bis(trifluoromethyl)quinolin-4-yl]-[(2S)-piperidin-2-yl]methanol, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Lin, J.Q, El Sahili, A, Lescar, J. | | Deposit date: | 2020-11-18 | | Release date: | 2021-12-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Identification of an inhibitory pocket in falcilysin provides a new avenue for malaria drug development.
Cell Chem Biol, 31, 2024
|
|
6I7W
 
 | | Structure of the periplasmic binding protein (PBP) AccA in complex with 2-glucose-2-O-lactic acid phosphate (G2LP) from Agrobacterium fabrum C58 | | Descriptor: | 2-O-[(R)-{[(2S)-1,1-dihydroxypropan-2-yl]oxy}(hydroxy)phosphoryl]-alpha-D-glucopyranose, 2-O-[(R)-{[(2S)-1,1-dihydroxypropan-2-yl]oxy}(hydroxy)phosphoryl]-beta-D-glucopyranose, ABC transporter, ... | | Authors: | Morera, S, Vigouroux, A, El Sahili, A. | | Deposit date: | 2018-11-19 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthesis of a non-natural glucose-2-phosphate ester able to dupe the acc system of Agrobacterium fabrum.
Org. Biomol. Chem., 17, 2019
|
|
5FSW
 
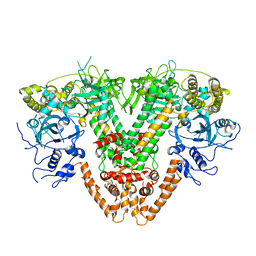 | | RNA dependent RNA polymerase QDE-1 from Thielavia terrestris | | Descriptor: | RNA DEPENDENT RNA POLYMERASE QDE-1 | | Authors: | Qian, X, Hamid, F.M, El Sahili, A, Darwis, D.A, Wong, Y.H, Bhushan, S, Makeyev, E.V, Lescar, J. | | Deposit date: | 2016-01-08 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Functional Evolution in Orthologous Cell-Encoded RNA-Dependent RNA Polymerases
J.Biol.Chem., 291, 2016
|
|
6ITO
 
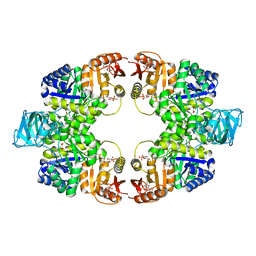 | | Crystal structure of pyruvate kinase (PYK) from Mycobacterium tuberculosis in complex with Oxalate, AMP and inhibitor Ribose 5-Phosphate | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, OXALATE ION, ... | | Authors: | Zhong, W, Cai, Q, El Sahili, A, Mu, Y, Lescar, J, Dedon, P.C. | | Deposit date: | 2018-11-24 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Pyruvate Kinase Regulates the Pentose-Phosphate Pathway in Response to Hypoxia in Mycobacterium tuberculosis.
J.Mol.Biol., 431, 2019
|
|
8CAW
 
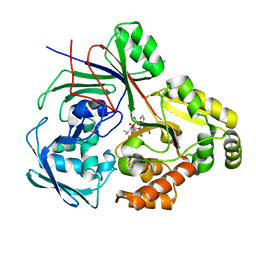 | | PBP AccA from A. tumefaciens Bo542 in complex with agrocin84 | | Descriptor: | 1,2-ETHANEDIOL, Agrocinopine utilization periplasmic binding protein AccA, [(2R,3R,4S,5S,6R)-6-(hydroxymethyl)-2,4,5-tris(oxidanyl)oxan-3-yl]oxy-N-[9-[(2R,3S,5R)-5-[[[(2R,3S)-4-methyl-2,3-bis(oxidanyl)pentanoyl]amino]-oxidanyl-phosphoryl]oxy-3-oxidanyl-oxolan-2-yl]purin-6-yl]phosphonamidic acid, ... | | Authors: | Morera, S, Vigouroux, A, El Sahili, A. | | Deposit date: | 2023-01-24 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.256 Å) | | Cite: | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
8HO4
 
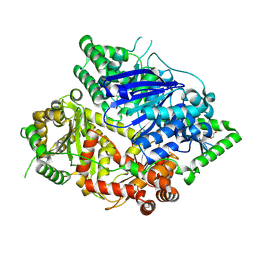 | | Falcilysin in complex with MMV000848 | | Descriptor: | (2R)-1-(9H-carbazol-9-yl)-3-(cyclopentylamino)propan-2-ol, 1,2-ETHANEDIOL, Falcilysin, ... | | Authors: | Lin, J.Q, Chung, Z, Lescar, J. | | Deposit date: | 2022-12-09 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Identification of an inhibitory pocket in falcilysin provides a new avenue for malaria drug development.
Cell Chem Biol, 31, 2024
|
|
8W7H
 
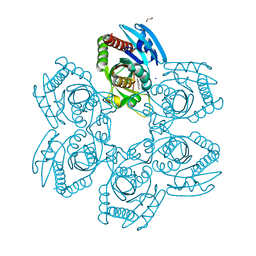 | | Purine Nucleoside Phosphorylase in complex with MMV000848 | | Descriptor: | (2R)-1-(9H-carbazol-9-yl)-3-(cyclopentylamino)propan-2-ol, 1,2-ETHANEDIOL, PHOSPHATE ION, ... | | Authors: | Chung, Z, Lin, J.Q, Lescar, J. | | Deposit date: | 2023-08-30 | | Release date: | 2024-01-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Identification and structural validation of purine nucleoside phosphorylase from Plasmodium falciparum as a target of MMV000848.
J.Biol.Chem., 300, 2023
|
|
6AFK
 
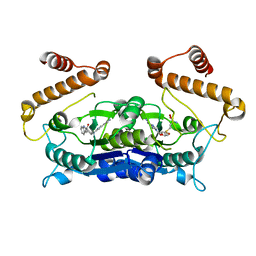 | | Crystal structure of TrmD from Pseudomonas aeruginosa in complex with active-site inhibitor | | Descriptor: | N-{(3S)-1-[3-(pyridin-4-yl)-1H-pyrazol-5-yl]piperidin-3-yl}-1H-indole-2-carboxamide, S-ADENOSYLMETHIONINE, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Zhong, W, Koay, A, Wong, Y.W, Sahili, A.E, Nah, Q, Kang, C, Poulsen, A, Chionh, Y.K, McBee, M, Matter, A, Hill, J, Lescar, J, Dedon, P.C. | | Deposit date: | 2018-08-08 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Targeting the Bacterial Epitranscriptome for Antibiotic Development: Discovery of Novel tRNA-(N1G37) Methyltransferase (TrmD) Inhibitors.
Acs Infect Dis., 5, 2019
|
|
5OT8
 
 | |
5OVZ
 
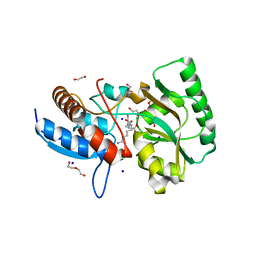 | | High resolution structure of the PBP NocT in complex with nopaline | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, N-[(1S)-4-carbamimidamido-1-carboxybutyl]-D-glutamic acid, ... | | Authors: | Vigouroux, A, Morera, S. | | Deposit date: | 2017-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Agrobacterium uses a unique ligand-binding mode for trapping opines and acquiring a competitive advantage in the niche construction on plant host.
Plos Pathog., 10, 2014
|
|
5OTA
 
 | |
5ORE
 
 | |
5OT9
 
 | |
5OTC
 
 | |
6U5O
 
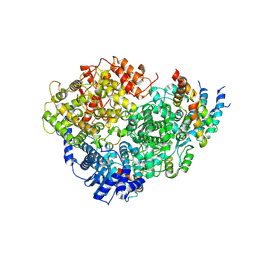 | | Structure of the Human Metapneumovirus Polymerase bound to the phosphoprotein tetramer | | Descriptor: | Phosphoprotein, RNA-directed RNA polymerase L | | Authors: | Pan, J, Qian, X, Lattmann, S, Sahili, A.E, Yeo, T.H, Kalocsay, M, Fearns, R, Lescar, J. | | Deposit date: | 2019-08-28 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of the human metapneumovirus polymerase phosphoprotein complex.
Nature, 577, 2020
|
|
5ORG
 
 | |
5K5M
 
 | |
7F5P
 
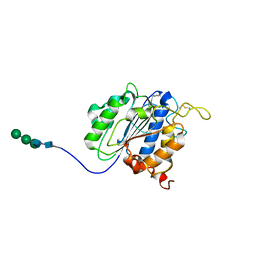 | | The crystal structure of VyPAL2-C214A, a dead mutant of VyPAL2 peptide asparaginyl ligase in form I | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Peptide Asparaginyl Ligases, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hu, S, Sahili, A, Lescar, J. | | Deposit date: | 2021-06-22 | | Release date: | 2022-06-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for proenzyme maturation, substrate recognition, and ligation by a hyperactive peptide asparaginyl ligase.
Plant Cell, 34, 2022
|
|
7F5Q
 
 | | The crystal structure of VyPAL2 peptide asparaginyl ligase in its active enzyme form | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hu, S, Sahili, A, Lescar, J. | | Deposit date: | 2021-06-22 | | Release date: | 2022-06-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for proenzyme maturation, substrate recognition, and ligation by a hyperactive peptide asparaginyl ligase.
Plant Cell, 34, 2022
|
|
