1K7L
 
 | | The 2.5 Angstrom resolution crystal structure of the human PPARalpha ligand binding domain bound with GW409544 and a co-activator peptide. | | Descriptor: | 2-(1-METHYL-3-OXO-3-PHENYL-PROPYLAMINO)-3-{4-[2-(5-METHYL-2-PHENYL-OXAZOL-4-YL)-ETHOXY]-PHENYL}-PROPIONIC ACID, Peroxisome proliferator activated receptor alpha, YTTRIUM (III) ION, ... | | Authors: | Xu, H.E, Lambert, M.H, Montana, V.G, Moore, L.B, Collins, J.L, Oplinger, J.A, Kliewer, S.A, Gampe Jr, R.T, McKee, D.D, Moore, J.T, Willson, T.M. | | Deposit date: | 2001-10-19 | | Release date: | 2001-12-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural determinants of ligand binding selectivity between the peroxisome proliferator-activated receptors.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
4PIK
 
 | |
4PFR
 
 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM RHODOBACTER SPHAEROIDES (Rsph17029_3541, TARGET EFI-510203), APO OPEN PARTIALLY DISORDERED | | Descriptor: | D-MALATE, TRAP dicarboxylate transporter, DctP subunit | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-04-30 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
1K8G
 
 | |
1I90
 
 | | CARBONIC ANHYDRASE II COMPLEXED WITH AL-8520 2H-THIENO[3,2-E]-1,2-THIAZINE-6-SULFONAMIDE, 4-AMINO-3,4-DIHYDRO-2-(3-METHOXYPROPYL)-, 1,1-DIOXIDE, (R) | | Descriptor: | 4-AMINO-6-[N-(3-METHOXYLPROPYL)-2H-THIENO[3,2-E][1,2]THIAZINE 1,1-DIOXIDE]-SULFONAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Kim, C.-Y, Chang, J.S, Liao, J, May, J.A, Christianson, D.W. | | Deposit date: | 2001-03-16 | | Release date: | 2001-03-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural aspects of isozyme selectivity in the binding of inhibitors to carbonic anhydrases II and IV.
J.Med.Chem., 45, 2002
|
|
1I9X
 
 | |
1I8Z
 
 | | CARBONIC ANHYDRASE II COMPLEXED WITH AL-6629 2H-THIENO[3,2-E]-1,2-THIAZINE-6-SULFONAMIDE, 2-(3-METHOXYPHENYL)-3-(4-MORPHOLINYL)-, 1,1-DIOXIDE | | Descriptor: | 6-[N-(3-METHOXY-PHENYL)-3-(MORPHOLIN-4-YLMETHYL)-2H-THIENO[3,2-E]-1,2-THIAZINE-1,1,-DIOXIDE]-SULFONAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Kim, C.-Y, Chang, J.S, Liao, J, May, J.A, Christianson, D.W. | | Deposit date: | 2001-03-16 | | Release date: | 2001-03-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural aspects of isozyme selectivity in the binding of inhibitors to carbonic anhydrases II and IV.
J.Med.Chem., 45, 2002
|
|
4R2F
 
 | | Crystal structure of sugar transporter ACHL_0255 from Arthrobacter chlorophenolicus A6, target EFI-510633, with bound laminaribiose | | Descriptor: | Extracellular solute-binding protein family 1, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Siedel, R.D, Hillerich, B, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-08-11 | | Release date: | 2014-08-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of sugar transporter ACHL_0255 from Arthrobacter chlorophenolicus, target EFI-510633
To be Published
|
|
1I91
 
 | | CARBONIC ANHYDRASE II COMPLEXED WITH AL-6619 2H-THIENO[3,2-E]-1,2-THIAZINE-6-SULFONAMIDE, 2-(3-HYDROXYPHENYL)-3-(4-MORPHOLINYL)-, 1,1-DIOXIDE | | Descriptor: | 6-[N-(3-HYDROXY-PHENYL)-3-(MORPHOLIN-4-YLMETHYL)-2H-THIENO[3,2-E]-1,2-THIAZINE-1,1,-DIOXIDE]-SULFONAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Kim, C.-Y, Chang, J.S, Liao, J, May, J.A, Christianson, D.W. | | Deposit date: | 2001-03-16 | | Release date: | 2001-03-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural aspects of isozyme selectivity in the binding of inhibitors to carbonic anhydrases II and IV.
J.Med.Chem., 45, 2002
|
|
4R6Y
 
 | | Crystal structure of solute-binding protein stm0429 from salmonella enterica subsp. enterica serovar typhimurium str. lt2, target efi-510776, a closed conformation, in complex with glycerol and acetate | | Descriptor: | ACETATE ION, GLYCEROL, Putative 2-aminoethylphosphonate-binding periplasmic protein | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al obaidi, N, Chamala, S, Attonito, J.D, Scott glenn, A, Chowdhury, S, Lafleur, J, Siedel, R.D, Hillerich, B, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-08-26 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Crystal Structure of Transporter STM0429 from Salmonella Enterica
To be Published
|
|
4O7H
 
 | | Crystal structure of a glutathione S-transferase from Rhodospirillum rubrum F11, Target EFI-507460 | | Descriptor: | Glutathione S-transferase | | Authors: | Kim, J, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Stead, M, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Attonito, J.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-12-24 | | Release date: | 2014-01-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a glutathione S-transferase from Rhodospirillum rubrum F11, Target EFI-507460
TO BE PUBLISHED
|
|
1JVQ
 
 | | Crystal structure at 2.6A of the ternary complex between antithrombin, a P14-P8 reactive loop peptide, and an exogenous tetrapeptide | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ANTITHROMBIN-III, ... | | Authors: | Zhou, A, Huntington, J.A, Lomas, D.A, Carrell, R.W, Stein, P.E. | | Deposit date: | 2001-08-31 | | Release date: | 2003-06-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | How small peptides block and reverse serpin polymerisation
J.Mol.Biol., 342, 2004
|
|
1JB5
 
 | | CRYSTAL STRUCTURE OF NTF2 M118E MUTANT | | Descriptor: | NUCLEAR TRANSPORT FACTOR 2 | | Authors: | Chaillan-Huntington, C, Butler, P.J, Huntington, J.A, Akin, D, Feldherr, C, Stewart, M. | | Deposit date: | 2001-06-01 | | Release date: | 2002-03-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | NTF2 monomer-dimer equilibrium.
J.Mol.Biol., 314, 2001
|
|
4PCD
 
 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM ROSEOBACTER DENITRIFICANS OCh 114 (RD1_1052, TARGET EFI-510238) WITH BOUND L-GALACTONATE | | Descriptor: | C4-dicarboxylate transport system, substrate-binding protein, putative, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-04-15 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4PGK
 
 | | Insights into Substrate and Metal Binding from the Crystal Structure of Cyanobacterial Aldehyde Deformylating Oxygenase with Substrate Bound | | Descriptor: | 11-[2-(2-ethoxyethoxy)ethoxy]undecanal, Aldehyde decarbonylase, FE (III) ION | | Authors: | Buer, B.C, Paul, B, Das, D, Stuckey, J.A, Marsh, E.N.G. | | Deposit date: | 2014-05-02 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Insights into substrate and metal binding from the crystal structure of cyanobacterial aldehyde deformylating oxygenase with substrate bound.
Acs Chem.Biol., 9, 2014
|
|
4PKA
 
 | |
4P94
 
 | | The crystal structure of the soluble domain of Sulfolobus acidocaldarius FlaF (residues 35-164) | | Descriptor: | Conserved flagellar protein F, SODIUM ION | | Authors: | Tsai, C.-L, Arvai, A.S, Ishida, J.P, Tainer, J.A. | | Deposit date: | 2014-04-02 | | Release date: | 2015-04-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | FlaF Is a beta-Sandwich Protein that Anchors the Archaellum in the Archaeal Cell Envelope by Binding the S-Layer Protein.
Structure, 23, 2015
|
|
1JPD
 
 | | L-Ala-D/L-Glu Epimerase | | Descriptor: | L-Ala-D/L-Glu Epimerase | | Authors: | Gulick, A.M, Schmidt, D.M.Z, Gerlt, J.A, Rayment, I. | | Deposit date: | 2001-08-01 | | Release date: | 2001-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: crystal structures of the L-Ala-D/L-Glu epimerases from Escherichia coli and Bacillus subtilis.
Biochemistry, 40, 2001
|
|
4PAK
 
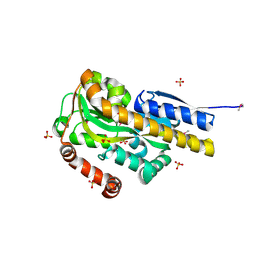 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM VERMINEPHROBACTER EISENIAE EF01-2 (Veis_3954, TARGET EFI-510324) A NEPHRIDIAL SYMBIONT OF THE EARTHWORM EISENIA FOETIDA, BOUND TO (R)-PANTOIC ACID | | Descriptor: | PANTOATE, SULFATE ION, TRAP dicarboxylate transporter, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-04-09 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4PQT
 
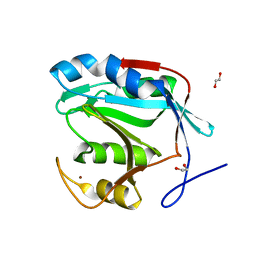 | | Insights into the mechanism of deubiquitination by JAMM deubiquitinases from co-crystal structures of enzyme with substrate and product | | Descriptor: | 1,2-ETHANEDIOL, AMSH-like protease sst2, Protein UBBP4, ... | | Authors: | Shrestha, R.K, Ronau, J.A, Das, C. | | Deposit date: | 2014-03-04 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Insights into the Mechanism of Deubiquitination by JAMM Deubiquitinases from Cocrystal Structures of the Enzyme with the Substrate and Product.
Biochemistry, 53, 2014
|
|
4PR6
 
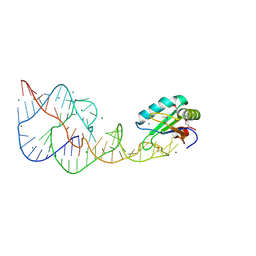 | | A Second Look at the HDV Ribozyme Structure and Dynamics. | | Descriptor: | HDV RIBOZYME SELF-CLEAVED, MAGNESIUM ION, U1 small nuclear ribonucleoprotein A | | Authors: | Kapral, G.J, Jain, S, Noeske, J, Doudna, J.A, Richardson, D.C, Richardson, J.S. | | Deposit date: | 2014-03-05 | | Release date: | 2014-10-29 | | Last modified: | 2014-11-26 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | New tools provide a second look at HDV ribozyme structure, dynamics and cleavage.
Nucleic Acids Res., 42, 2014
|
|
1KV8
 
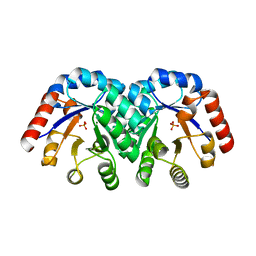 | | Crystal Structure of 3-Keto-L-Gulonate 6-Phosphate Decarboxylase | | Descriptor: | 3-Keto-L-Gulonate 6-Phosphate Decarboxylase, MAGNESIUM ION, PHOSPHATE ION | | Authors: | Wise, E, Yew, W.S, Babbitt, P.C, Gerlt, J.A, Rayment, I. | | Deposit date: | 2002-01-25 | | Release date: | 2002-04-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Homologous (beta/alpha)8-barrel enzymes that catalyze unrelated reactions: orotidine 5'-monophosphate decarboxylase and 3-keto-L-gulonate 6-phosphate decarboxylase.
Biochemistry, 41, 2002
|
|
4PC9
 
 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM ROSENBACTER DENITRIFICANS OCh 114 (RD1_1052, TARGET EFI-510238) WITH BOUND D-MANNONATE | | Descriptor: | C4-dicarboxylate transport system, substrate-binding protein, putative, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-04-14 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4PDH
 
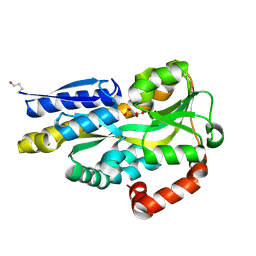 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM POLAROMONAS SP JS666 (Bpro_1871, TARGET EFI-510164) BOUND TO D-ERYTHRONATE | | Descriptor: | (2R,3R)-2,3,4-trihydroxybutanoic acid, TRAP dicarboxylate transporter, DctP subunit | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-04-18 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4PE3
 
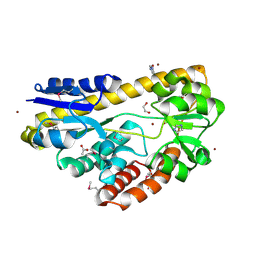 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM RHODOBACTER SPHAEROIDES (Rsph17029_3620, TARGET EFI-510199), APO OPEN STRUCTURE | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, IMIDAZOLE, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-04-22 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
