4QBS
 
 | |
6BB1
 
 | | Lactate Dehydrogenase in complex with inhibitor (R)-5-((2-chlorophenyl)thio)-6'-(4-fluorophenoxy)-4-hydroxy-2-(thiophen-3-yl)-2,3-dihydro-[2,2'-bipyridin]-6(1H)-one | | Descriptor: | (2R)-5-[(2-chlorophenyl)sulfanyl]-6'-(4-fluorophenoxy)-4-hydroxy-2-(thiophen-3-yl)-2,3-dihydro[2,2'-bipyridin]-6(1H)-one, L-lactate dehydrogenase A chain, LACTIC ACID, ... | | Authors: | Ultsch, M, Eigenbrot, C. | | Deposit date: | 2017-10-16 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-guided optimization and in vivo activities of hydroxylactone and hydroxylactam Inhibitors of Human Lactate Dehydrogenase
To Be Published
|
|
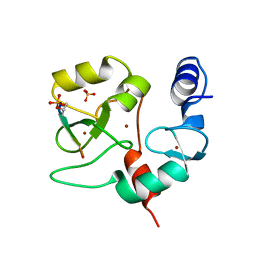
6LAN
 
 | | Structure of CCDC50 and LC3B complex | | Descriptor: | Coiled-coil domain-containing protein 50,Microtubule-associated proteins 1A/1B light chain 3B | | Authors: | Liu, L, Li, J, Hou, P. | | Deposit date: | 2019-11-12 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | A novel selective autophagy receptor, CCDC50, delivers K63 polyubiquitination-activated RIG-I/MDA5 for degradation during viral infection.
Cell Res., 31, 2021
|
|
6BB2
 
 | | Lactate Dehydrogenase in complex with inhibitor (S)-5-((2-chlorophenyl)thio)-6'-(4-fluorophenoxy)-4-hydroxy-2-(thiophen-3-yl)-2,3-dihydro-[2,2'-bipyridin]-6(1H)-one | | Descriptor: | (2S)-5-[(2-chlorophenyl)sulfanyl]-6'-(4-fluorophenoxy)-4-hydroxy-2-(thiophen-3-yl)-2,3-dihydro[2,2'-bipyridin]-6(1H)-one, L-lactate dehydrogenase A chain, LACTIC ACID, ... | | Authors: | Ultsch, M, Eigenbrot, C. | | Deposit date: | 2017-10-16 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structure-guided optimization and in vivo activities of hydroxylactone and hydroxylactam Inhibitors of Human Lactate Dehydrogenase
To Be Published
|
|
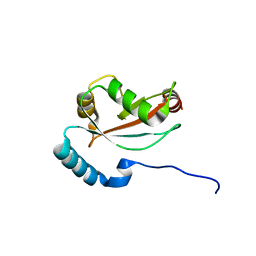
6BAG
 
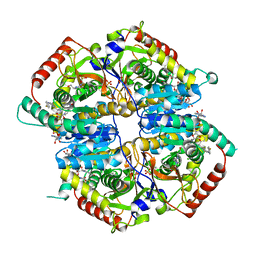 | | Lactate Dehydrogenase in complex with inhibitor (R)-5-((2-chlorophenyl)thio)-6'-((4-fluorophenyl)amino)-4-hydroxy-2-(thiophen-3-yl)-2,3-dihydro-[2,2'-bipyridin]-6(1H)-one | | Descriptor: | (2R)-5-[(2-chlorophenyl)sulfanyl]-6'-[(4-fluorophenyl)amino]-4-hydroxy-2-(thiophen-3-yl)-2,3-dihydro[2,2'-bipyridin]-6(1H)-one, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, L-lactate dehydrogenase A chain, ... | | Authors: | Ultsch, M, Eigenbrot, C. | | Deposit date: | 2017-10-12 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-guided optimization and in vivo activities of hydroxylactone and hydroxylactam Inhibitors of Human Lactate Dehydrogenase
To Be Published
|
|
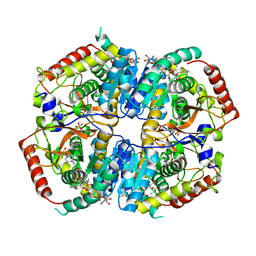
6BB0
 
 | |
6BAZ
 
 | | Lactate Dehydrogenase in complex with inhibitor (S)-5-((2-chlorophenyl)thio)-6'-((4-fluorophenyl)amino)-4-hydroxy-2-(thiophen-3-yl)-2,3-dihydro-[2,2'-bipyridin]-6(1H)-one | | Descriptor: | (3S,6S)-3-[(2-chlorophenyl)sulfanyl]-6-{6-[(4-fluorophenyl)amino]pyridin-2-yl}-6-(thiophen-3-yl)piperidine-2,4-dione, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, L-lactate dehydrogenase A chain, ... | | Authors: | Ultsch, M, Eigenbrot, C. | | Deposit date: | 2017-10-16 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-guided optimization and in vivo activities of hydroxylactone and hydroxylactam Inhibitors of Human Lactate Dehydrogenase
To Be Published
|
|
6BAD
 
 | |
5Y8Y
 
 | | Crystal Structure Analysis of the BRD4 | | Descriptor: | 1,2-ETHANEDIOL, 5-bromanyl-2-methoxy-N-(6-methoxy-3-methyl-1,2-benzoxazol-5-yl)benzenesulfonamide, Bromodomain-containing protein 4, ... | | Authors: | Xu, Y, Zhang, Y, Song, M, Wang, C. | | Deposit date: | 2017-08-21 | | Release date: | 2018-06-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure-Based Discovery and Optimization of Benzo[ d]isoxazole Derivatives as Potent and Selective BET Inhibitors for Potential Treatment of Castration-Resistant Prostate Cancer (CRPC)
J. Med. Chem., 61, 2018
|
|
5Y8Z
 
 | | Crystal Structure Analysis of the BRD4 | | Descriptor: | 1,2-ETHANEDIOL, 5-bromanyl-N-(3,6-dimethyl-1,2-benzoxazol-5-yl)-2-methoxy-benzenesulfonamide, Bromodomain-containing protein 4, ... | | Authors: | Xu, Y, Zhang, Y, Song, M, Wang, C. | | Deposit date: | 2017-08-22 | | Release date: | 2018-06-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structure-Based Discovery and Optimization of Benzo[ d]isoxazole Derivatives as Potent and Selective BET Inhibitors for Potential Treatment of Castration-Resistant Prostate Cancer (CRPC)
J. Med. Chem., 61, 2018
|
|
5Y8C
 
 | | Crystal Structure Analysis of the BRD4 | | Descriptor: | 1,2-ETHANEDIOL, 5-chloranyl-2-methoxy-N-(6-methoxy-3-methyl-1,2-benzoxazol-5-yl)benzenesulfonamide, Bromodomain-containing protein 4, ... | | Authors: | Xu, Y, Zhang, Y, Song, M, Wang, C. | | Deposit date: | 2017-08-21 | | Release date: | 2018-06-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structure-Based Discovery and Optimization of Benzo[ d]isoxazole Derivatives as Potent and Selective BET Inhibitors for Potential Treatment of Castration-Resistant Prostate Cancer (CRPC)
J. Med. Chem., 61, 2018
|
|
3L9M
 
 | | Crystal structure of PKAB3 (pka triple mutant V123A, L173M, Q181K) with compound 18 | | Descriptor: | (2S)-N~1~-[5-(3-methyl-1H-indazol-5-yl)-1,3,4-thiadiazol-2-yl]-3-(4-methylphenyl)propane-1,2-diamine, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Huang, X. | | Deposit date: | 2010-01-05 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Azole-based inhibitors of AKT/PKB for the treatment of cancer.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2Q2Y
 
 | | Crystal Structure of KSP in complex with Inhibitor 1 | | Descriptor: | 1-{(3R,3AR)-3-[3-(4-ACETYLPIPERAZIN-1-YL)PROPYL]-8-FLUORO-3-PHENYL-3A,4-DIHYDRO-3H-PYRAZOLO[5,1-C][1,4]BENZOXAZIN-2-YL}ETHANONE, ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF11, ... | | Authors: | Yan, Y. | | Deposit date: | 2007-05-29 | | Release date: | 2007-09-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Kinesin spindle protein (KSP) inhibitors. Part 8: Design and synthesis of 1,4-diaryl-4,5-dihydropyrazoles as potent inhibitors of the mitotic kinesin KSP.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
3L9N
 
 | | crystal structure of PKAB3 (pka triple mutant V123A, L173M, Q181K) with compound 27 | | Descriptor: | (2S)-N~1~-[5-(1H-indazol-5-yl)-1,3,4-thiadiazol-2-yl]-3-(4-methylphenyl)propane-1,2-diamine, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Huang, X. | | Deposit date: | 2010-01-05 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Azole-based inhibitors of AKT/PKB for the treatment of cancer.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3L9L
 
 | | Crystal structure of pka with compound 36 | | Descriptor: | 5-[2-({(2S)-2-amino-3-[4-(trifluoromethyl)phenyl]propyl}amino)-1,3-thiazol-5-yl]-1,3-dihydro-2H-indol-2-one, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Huang, X. | | Deposit date: | 2010-01-05 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Azole-based inhibitors of AKT/PKB for the treatment of cancer.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
4QBR
 
 | |
5Y93
 
 | | Crystal Structure Analysis of the BRD4 | | Descriptor: | 1,2-ETHANEDIOL, 2-[[5-[(5-bromanyl-2-methoxy-phenyl)sulfonylamino]-3-methyl-1,2-benzoxazol-6-yl]oxy]-N-(2-morpholin-4-ylethyl)ethanamide, Bromodomain-containing protein 4, ... | | Authors: | Xu, Y, Zhang, Y, Song, M, Wang, C. | | Deposit date: | 2017-08-22 | | Release date: | 2018-06-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure-Based Discovery and Optimization of Benzo[ d]isoxazole Derivatives as Potent and Selective BET Inhibitors for Potential Treatment of Castration-Resistant Prostate Cancer (CRPC)
J. Med. Chem., 61, 2018
|
|
2Q2Z
 
 | | Crystal Structure of KSP in Complex with Inhibitor 22 | | Descriptor: | 1-[(4R)-4-[3-(4-ACETYLPIPERAZIN-1-YL)PROPYL]-1-(2-FLUORO-5-METHYLPHENYL)-4-PHENYL-4,5-DIHYDRO-1H-PYRAZOL-3-YL]ETHANONE, ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF11, ... | | Authors: | Yan, Y. | | Deposit date: | 2007-05-29 | | Release date: | 2007-09-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Kinesin spindle protein (KSP) inhibitors. Part 8: Design and synthesis of 1,4-diaryl-4,5-dihydropyrazoles as potent inhibitors of the mitotic kinesin KSP.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
7CSW
 
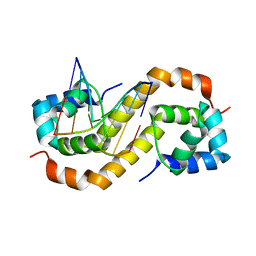 | | Pseudomonas aeruginosa antitoxin HigA with pa2440 promoter | | Descriptor: | HTH cro/C1-type domain-containing protein, pa2440 | | Authors: | Song, Y.J, Luo, G.H, Bao, R. | | Deposit date: | 2020-08-17 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Pseudomonas aeruginosa antitoxin HigA functions as a diverse regulatory factor by recognizing specific pseudopalindromic DNA motifs.
Environ.Microbiol., 23, 2021
|
|
7CSV
 
 | | Pseudomonas aeruginosa antitoxin HigA | | Descriptor: | HTH cro/C1-type domain-containing protein | | Authors: | Song, Y.J, Luo, G.H, Bao, R. | | Deposit date: | 2020-08-17 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Pseudomonas aeruginosa antitoxin HigA functions as a diverse regulatory factor by recognizing specific pseudopalindromic DNA motifs.
Environ.Microbiol., 23, 2021
|
|
7CSY
 
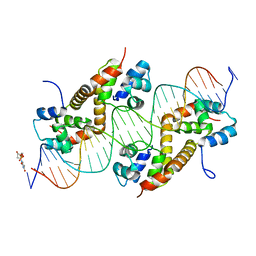 | | Pseudomonas aeruginosa antitoxin HigA with higBA promoter | | Descriptor: | DNA (28-MER), DNA (29-MER), HTH cro/C1-type domain-containing protein, ... | | Authors: | Song, Y.J, Luo, G.H, Bao, R. | | Deposit date: | 2020-08-17 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Pseudomonas aeruginosa antitoxin HigA functions as a diverse regulatory factor by recognizing specific pseudopalindromic DNA motifs.
Environ.Microbiol., 23, 2021
|
|
6IUO
 
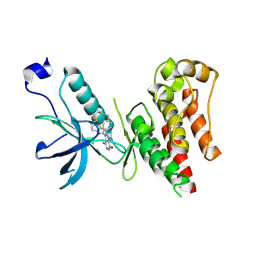 | | Crystal structure of FGFR4 kinase domain in complex with a covalent inhibitor | | Descriptor: | Fibroblast growth factor receptor 4, N-({4-[4-amino-3-(3,5-dimethyl-1-benzofuran-2-yl)-7-oxo-6,7-dihydro-2H-pyrazolo[3,4-d]pyridazin-2-yl]phenyl}methyl)prop-2-enamide | | Authors: | Xu, Y, Liu, Q. | | Deposit date: | 2018-11-29 | | Release date: | 2019-10-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery and Development of a Series of Pyrazolo[3,4-d]pyridazinone Compounds as the Novel Covalent Fibroblast Growth Factor Receptor Inhibitors by the Rational Drug Design.
J.Med.Chem., 62, 2019
|
|
6ITJ
 
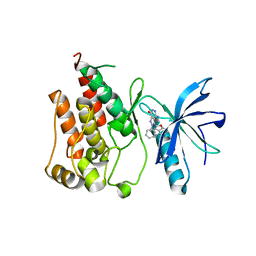 | | Crystal structure of FGFR1 kinase domain in complex with compound 3 | | Descriptor: | 4-azanyl-3-(3,5-dimethyl-1-benzofuran-2-yl)-2-phenyl-6~{H}-pyrazolo[3,4-d]pyridazin-7-one, Fibroblast growth factor receptor 1 | | Authors: | Xu, Y, Liu, Q. | | Deposit date: | 2018-11-23 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.994 Å) | | Cite: | Discovery and Development of a Series of Pyrazolo[3,4-d]pyridazinone Compounds as the Novel Covalent Fibroblast Growth Factor Receptor Inhibitors by the Rational Drug Design.
J.Med.Chem., 62, 2019
|
|
3ZDU
 
 | | Crystal structure of the human CDKL3 kinase domain | | Descriptor: | 1,2-ETHANEDIOL, CYCLIN-DEPENDENT KINASE-LIKE 3, SODIUM ION, ... | | Authors: | Canning, P, Elkins, J.M, Goubin, S, Mahajan, P, Pike, A.C.W, Quigley, A, MacKenzie, A, Carpenter, E.P, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2012-11-30 | | Release date: | 2013-03-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | CDKL Family Kinases Have Evolved Distinct Structural Features and Ciliary Function.
Cell Rep, 22, 2018
|
|
6CHM
 
 | |
