5XQI
 
 | | Crystal structure of full-length human Rogdi | | Descriptor: | Protein rogdi homolog | | Authors: | Lee, H, Lee, C. | | Deposit date: | 2017-06-07 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of human Rogdi provides insight into the causes of Kohlschutter-Tonz Syndrome
Sci Rep, 7, 2017
|
|
5H7B
 
 | | Crystal structure of a repeat protein with five Protein A repeat modules | | Descriptor: | Immunoglobulin G-binding protein A | | Authors: | Youn, S.J, Kwon, N.Y, Lee, J.H, Kim, J.H, Lee, H, Lee, J.O. | | Deposit date: | 2016-11-17 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Construction of novel repeat proteins with rigid and predictable structures using a shared helix method.
Sci Rep, 7, 2017
|
|
5H79
 
 | | Crystal structure of a repeat protein with three Protein A repeat module | | Descriptor: | Immunoglobulin G-binding protein A | | Authors: | Youn, S.J, Kwon, N.Y, Lee, J.H, Kim, J.H, Lee, H, Lee, J.O. | | Deposit date: | 2016-11-17 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Construction of novel repeat proteins with rigid and predictable structures using a shared helix method.
Sci Rep, 7, 2017
|
|
5H75
 
 | | Crystal structure of the MrsD-Protein A fusion protein | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Mersacidin decarboxylase,Immunoglobulin G-binding protein A | | Authors: | Youn, S.J, Kwon, N.Y, Lee, J.H, Kim, J.H, Lee, H, Lee, J.O. | | Deposit date: | 2016-11-17 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.738 Å) | | Cite: | Construction of novel repeat proteins with rigid and predictable structures using a shared helix method.
Sci Rep, 7, 2017
|
|
5H7D
 
 | | Crystal structure of the YgjG-protein A-Zpa963-calmodulin complex | | Descriptor: | CALCIUM ION, Putrescine aminotransferase,Immunoglobulin G-binding protein A, Zpa963,Calmodulin | | Authors: | Youn, S.J, Kwon, N.Y, Lee, J.H, Kim, J.H, Lee, H, Lee, J.O. | | Deposit date: | 2016-11-17 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Construction of novel repeat proteins with rigid and predictable structures using a shared helix method.
Sci Rep, 7, 2017
|
|
5H7A
 
 | | Crystal structure of a repeat protein with four Protein A repeat module | | Descriptor: | Immunoglobulin G-binding protein A | | Authors: | Youn, S.J, Kwon, N.Y, Lee, J.H, Kim, J.H, Lee, H, Lee, J.O. | | Deposit date: | 2016-11-17 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Construction of novel repeat proteins with rigid and predictable structures using a shared helix method.
Sci Rep, 7, 2017
|
|
5H76
 
 | | Crystal structure of the DARPin-Protein A fusion protein | | Descriptor: | DARPin,Immunoglobulin G-binding protein A | | Authors: | Youn, S.J, Kwon, N.Y, Lee, J.H, Kim, J.H, Lee, H, Lee, J.O. | | Deposit date: | 2016-11-17 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Construction of novel repeat proteins with rigid and predictable structures using a shared helix method.
Sci Rep, 7, 2017
|
|
5W4U
 
 | |
1X32
 
 | |
3FAP
 
 | |
6VOS
 
 | | Crystal structure of macaque anti-HIV-1 antibody RM20J | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, HEXAETHYLENE GLYCOL, NONAETHYLENE GLYCOL, ... | | Authors: | Yuan, M, Wilson, I.A. | | Deposit date: | 2020-01-31 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Mapping the immunogenic landscape of near-native HIV-1 envelope trimers in non-human primates.
Plos Pathog., 16, 2020
|
|
4EEY
 
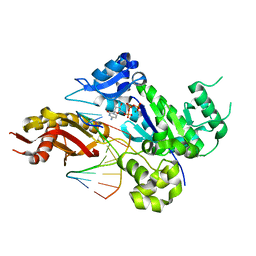 | | Crystal structure of human DNA polymerase eta in ternary complex with a cisplatin DNA adduct | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 5'-D(*CP*TP*TP*GP*GP*TP*CP*TP*CP*CP*TP*CP*C)-3', 5'-D(*TP*GP*GP*AP*GP*GP*AP*GP*A)-3', ... | | Authors: | Ummat, A, Rechkoblit, O, Jain, R, Choudhury, J.R, Johnson, R.E, Silverstein, T.D, Buku, A, Lone, S, Prakash, L, Prakash, S, Aggarwal, A.K. | | Deposit date: | 2012-03-28 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural basis for cisplatin DNA damage tolerance by human polymerase {eta} during cancer chemotherapy.
Nat.Struct.Mol.Biol., 19, 2012
|
|
6WB0
 
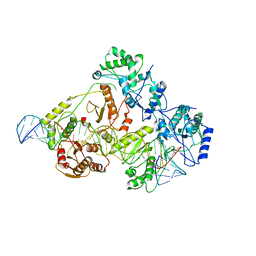 | | +3 extended HIV-1 reverse transcriptase initiation complex core (pre-translocation state) | | Descriptor: | HIV-1 viral RNA genome fragment, Reverse transcriptase/ribonuclease H, reverse transcriptase p51 subunit, ... | | Authors: | Larsen, K.P, Jackson, L.N, Kappel, K, Zhang, J, Puglisi, E.V. | | Deposit date: | 2020-03-26 | | Release date: | 2020-06-24 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Distinct Conformational States Underlie Pausing during Initiation of HIV-1 Reverse Transcription.
J.Mol.Biol., 432, 2020
|
|
6VOR
 
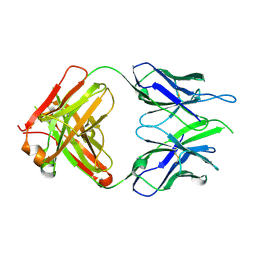 | | Crystal structure of macaque anti-HIV-1 antibody RM20E1 | | Descriptor: | GLYCINE, RM20E1 Fab heavy chain, RM20E1 Fab light chain | | Authors: | Yuan, M, Wilson, I.A. | | Deposit date: | 2020-01-31 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mapping the immunogenic landscape of near-native HIV-1 envelope trimers in non-human primates.
Plos Pathog., 16, 2020
|
|
6WAZ
 
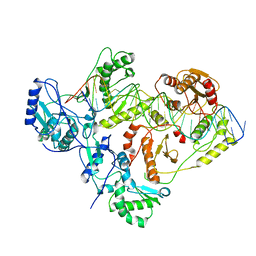 | | +1 extended HIV-1 reverse transcriptase initiation complex core (pre-translocation state) | | Descriptor: | HIV-1 viral RNA genome fragment, Reverse transcriptase p51 subunit, Reverse transcriptase/ribonuclease H, ... | | Authors: | Larsen, K.P, Jackson, L.N, Kappel, K, Zhang, J, Puglisi, E.V. | | Deposit date: | 2020-03-26 | | Release date: | 2020-06-24 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Distinct Conformational States Underlie Pausing during Initiation of HIV-1 Reverse Transcription.
J.Mol.Biol., 432, 2020
|
|
6WB1
 
 | | +3 extended HIV-1 reverse transcriptase initiation complex core (intermediate state) | | Descriptor: | HIV-1 viral RNA genome fragment, REVERSE TRANSCRIPTASE/RIBONUCLEASE H, reverse transcriptase p51 subunit, ... | | Authors: | Larsen, K.P, Jackson, L.N, Kappel, K, Zhang, J, Chen, D.H, Puglisi, E.V. | | Deposit date: | 2020-03-26 | | Release date: | 2020-06-24 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Distinct Conformational States Underlie Pausing during Initiation of HIV-1 Reverse Transcription.
J.Mol.Biol., 432, 2020
|
|
6VSR
 
 | |
6WB2
 
 | | +3 extended HIV-1 reverse transcriptase initiation complex core (displaced state) | | Descriptor: | HIV-1 viral RNA genome fragment, Reverse transcriptase/ribonuclease H, reverse transcriptase p51 subunit, ... | | Authors: | Larsen, K.P, Jackson, L.N, Kappel, K, Zhang, J, Chen, D.H, Puglisi, E.V. | | Deposit date: | 2020-03-26 | | Release date: | 2020-06-24 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Distinct Conformational States Underlie Pausing during Initiation of HIV-1 Reverse Transcription.
J.Mol.Biol., 432, 2020
|
|
4LWA
 
 | | Structure of Bacillus subtilis nitric oxide synthase in complex with ((2S, 3S)-1,3-bis((6-(2,5-dimethyl-1H-pyrrol-1-yl)-4-methylpyridin-2-yl)methoxy)-2-aminobutane | | Descriptor: | 6,6'-{[(2S,3S)-2-aminobutane-1,3-diyl]bis(oxymethanediyl)}bis(4-methylpyridin-2-amine), CHLORIDE ION, GLYCEROL, ... | | Authors: | Holden, J.K, Li, H, Poulos, T.P. | | Deposit date: | 2013-07-26 | | Release date: | 2013-10-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural and biological studies on bacterial nitric oxide synthase inhibitors.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
7MP7
 
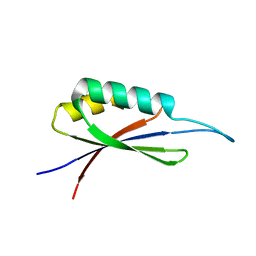 | | Rules for designing protein fold switches and their implications for the folding code | | Descriptor: | Sb3 | | Authors: | He, Y, Chen, Y, Ruan, B, Choi, J, Chen, Y, Motabar, D, Solomon, T, Simmerman, R, Kauffman, T, Gallagher, T, Bryan, P, Orban, J. | | Deposit date: | 2021-05-04 | | Release date: | 2022-05-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Design and characterization of a protein fold switching network.
Nat Commun, 14, 2023
|
|
7MN1
 
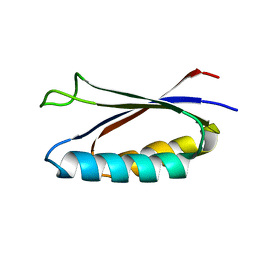 | | Rules for designing protein fold switches and their implications for the folding code | | Descriptor: | Sa1 | | Authors: | He, Y, Chen, Y, Ruan, B, Choi, J, Chen, Y, Motabar, D, Solomon, T, Simmerman, R, Kauffman, T, Gallagher, T, Bryan, P, Orban, J. | | Deposit date: | 2021-04-30 | | Release date: | 2022-05-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Design and characterization of a protein fold switching network.
Nat Commun, 14, 2023
|
|
7MQ4
 
 | | Rules for designing protein fold switches and their implications for the folding code | | Descriptor: | Sb1 | | Authors: | He, Y, Chen, Y, Ruan, B, Choi, J, Chen, Y, Motabar, D, Solomon, T, Simmerman, R, Kauffman, T, Gallagher, T, Bryan, P, Orban, J. | | Deposit date: | 2021-05-05 | | Release date: | 2022-05-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Design and characterization of a protein fold switching network.
Nat Commun, 14, 2023
|
|
6M2D
 
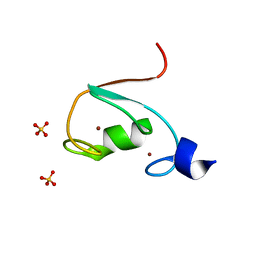 | | MUL1-RING domain | | Descriptor: | Mitochondrial ubiquitin ligase activator of NFKB 1, SULFATE ION, ZINC ION | | Authors: | Lee, S.O, Ryu, K.S, Chi, S.-W. | | Deposit date: | 2020-02-27 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.795 Å) | | Cite: | MUL1-RING recruits the substrate, p53-TAD as a complex with UBE2D2-UB conjugate.
Febs J., 2022
|
|
6M2C
 
 | |
4LWB
 
 | | Structure of Bacillus subtilis nitric oxide synthase in complex with 6-((((3R,5S)-5-(((6-amino-4-methylpyridin-2-yl)methoxy)methyl)pyrrolidin-3-yl)oxy)methyl)-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-({[(3R,5S)-5-{[(6-amino-4-methylpyridin-2-yl)methoxy]methyl}pyrrolidin-3-yl]oxy}methyl)-4-methylpyridin-2-amine, CHLORIDE ION, ... | | Authors: | Holden, J.K, Li, H, Poulos, T.L. | | Deposit date: | 2013-07-26 | | Release date: | 2013-10-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and biological studies on bacterial nitric oxide synthase inhibitors.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
