7KO0
 
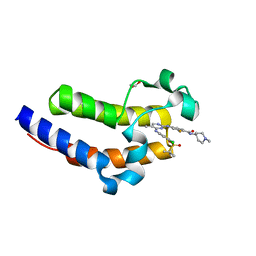 | | Crystal structure of the second bromodomain (BD2) of human BRD4 bound to SG3-179 | | Descriptor: | 1,2-ETHANEDIOL, 4-[[4-[[3-(~{tert}-butylsulfonylamino)-4-chloranyl-phenyl]amino]-5-methyl-pyrimidin-2-yl]amino]-2-fluoranyl-~{N}-(1-methylpiperidin-4-yl)benzamide, Bromodomain-containing protein 4 | | Authors: | Karim, M.R, Zhu, J.Y, Schonbrunn, E. | | Deposit date: | 2020-11-06 | | Release date: | 2021-10-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Differential BET Bromodomain Inhibition by Dihydropteridinone and Pyrimidodiazepinone Kinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
7L72
 
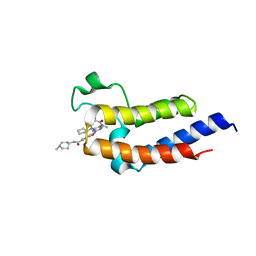 | | Crystal structure of the second bromodomain (BD2) of human BRD3 bound to Ro3280 | | Descriptor: | 4-[(9-cyclopentyl-7,7-difluoro-5-methyl-6-oxo-6,7,8,9-tetrahydro-5H-pyrimido[4,5-b][1,4]diazepin-2-yl)amino]-3-methoxy-N-(1-methylpiperidin-4-yl)benzamide, Bromodomain-containing protein 3 | | Authors: | Karim, M.R, Bikowitz, M, Schonbrunn, E. | | Deposit date: | 2020-12-25 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Differential BET Bromodomain Inhibition by Dihydropteridinone and Pyrimidodiazepinone Kinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
2V9Z
 
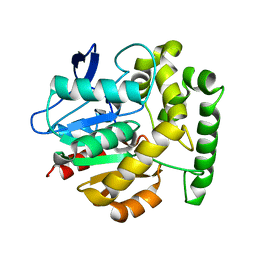 | | Structure of the Rhodococcus haloalkane dehalogenase mutant with enhanced enantioselectivity | | Descriptor: | HALOALKANE DEHALOGENASE | | Authors: | Koudelakova, T, Prokop, Z, Sato, Y, Lapkouski, M, Chovancova, E, Monincova, M, Jesenska, A, Emmer, J, Senda, T, Nagata, Y, Kuta Smatanova, I, Damborsky, J. | | Deposit date: | 2007-08-28 | | Release date: | 2008-09-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Rational Engineering of Rhodococcus Haloalkane Dehalogenase with Enhanced Enantioselectivity
To be Published
|
|
7ERQ
 
 | |
7ERP
 
 | |
7FDF
 
 | |
5UM3
 
 | |
5XEQ
 
 | | Crystal Structure of human MDGA1 and human neuroligin-2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MAM domain-containing glycosylphosphatidylinositol anchor protein 1, ... | | Authors: | Kim, H.M, Kim, J.A, Kim, D. | | Deposit date: | 2017-04-05 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.136 Å) | | Cite: | Structural Insights into Modulation of Neurexin-Neuroligin Trans-synaptic Adhesion by MDGA1/Neuroligin-2 Complex
Neuron, 94, 2017
|
|
5TDA
 
 | |
5TDB
 
 | | Crystal structure of the human UBR-box domain from UBR2 in complex with asymmetrically double methylated arginine peptide | | Descriptor: | 1,2-ETHANEDIOL, DA2-ILE-PHE-SER peptide, E3 ubiquitin-protein ligase UBR2, ... | | Authors: | Munoz-Escobar, J, Kozlov, G, Gehring, K. | | Deposit date: | 2016-09-19 | | Release date: | 2017-03-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.101 Å) | | Cite: | Bound Waters Mediate Binding of Diverse Substrates to a Ubiquitin Ligase.
Structure, 25, 2017
|
|
5TDD
 
 | | Human UBR-box from UBR2 in complex with HIFS peptide | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase UBR2, HIS-ILE-PHE-SER peptide, ... | | Authors: | Munoz-Escobar, J, Kozlov, G, Gehring, K. | | Deposit date: | 2016-09-19 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Bound Waters Mediate Binding of Diverse Substrates to a Ubiquitin Ligase.
Structure, 25, 2017
|
|
7LRQ
 
 | | Crystal structure of human SFPQ/NONO heterodimer, conserved DBHS region | | Descriptor: | CHLORIDE ION, Non-POU domain-containing octamer-binding protein, Splicing factor, ... | | Authors: | Marshall, A.C, Bond, C.S, Mohnen, I. | | Deposit date: | 2021-02-17 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Paraspeckle subnuclear bodies depend on dynamic heterodimerisation of DBHS RNA-binding proteins via their structured domains.
J.Biol.Chem., 298, 2022
|
|
5OWH
 
 | | High salt structure of human protein kinase CK2alpha in complex with 3-aminopropyl-4,5,6,7-tetrabromobenzimidazol | | Descriptor: | 3-[4,5,6,7-tetrakis(bromanyl)benzimidazol-1-yl]propan-1-amine, CHLORIDE ION, Casein kinase II subunit alpha | | Authors: | Niefind, K, Bretner, M, Chojnacki, C. | | Deposit date: | 2017-09-01 | | Release date: | 2018-07-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Biological properties and structural study of new aminoalkyl derivatives of benzimidazole and benzotriazole, dual inhibitors of CK2 and PIM1 kinases.
Bioorg. Chem., 80, 2018
|
|
5OWL
 
 | | Low salt structure of human protein kinase CK2alpha in complex with 3-aminopropyl-4,5,6,7-tetrabromobenzimidazol | | Descriptor: | 3-[4,5,6,7-tetrakis(bromanyl)benzimidazol-1-yl]propan-1-amine, Casein kinase II subunit alpha, GLYCEROL, ... | | Authors: | Niefind, K, Bretner, M, Chojnacki, C. | | Deposit date: | 2017-09-01 | | Release date: | 2018-07-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Biological properties and structural study of new aminoalkyl derivatives of benzimidazole and benzotriazole, dual inhibitors of CK2 and PIM1 kinases.
Bioorg. Chem., 80, 2018
|
|
8CC4
 
 | | LasB bound to phosphonic acid based inhibitor | | Descriptor: | CALCIUM ION, Elastase, ZINC ION, ... | | Authors: | Mueller, R, Sikandar, A. | | Deposit date: | 2023-01-26 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Inhibitors of the Elastase LasB for the Treatment of Pseudomonas aeruginosa Lung Infections.
Acs Cent.Sci., 9, 2023
|
|
6KQT
 
 | | Crystal Structure of GH136 lacto-N-biosidase from Eubacterium ramulus - native protein | | Descriptor: | SODIUM ION, TRIETHYLENE GLYCOL, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yamada, C, Arakawa, T, Pichler, M.J, Abou Hachem, M, Fushinobu, S. | | Deposit date: | 2019-08-18 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Butyrate producing colonic Clostridiales metabolise human milk oligosaccharides and cross feed on mucin via conserved pathways.
Nat Commun, 11, 2020
|
|
6KQS
 
 | | Crystal Structure of GH136 lacto-N-biosidase from Eubacterium ramulus - selenomethionine derivative | | Descriptor: | GLYCEROL, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, lacto-N-biosidase | | Authors: | Yamada, C, Arakawa, T, Pichler, M.J, Abou Hachem, M, Fushinobu, S. | | Deposit date: | 2019-08-18 | | Release date: | 2020-06-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Butyrate producing colonic Clostridiales metabolise human milk oligosaccharides and cross feed on mucin via conserved pathways.
Nat Commun, 11, 2020
|
|
7ODU
 
 | | Natural killer cell receptor NKR-P1B from Rattus norvegicus in complex with its cognate ligand Clr-11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C-type lectin domain family 2 member D11, ... | | Authors: | Skalova, T, Blaha, J, Kalouskova, B, Skorepa, O, Vanek, O, Dohnalek, J. | | Deposit date: | 2021-04-30 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Natural killer cell receptor NKR-P1B from Rattus norvegicus in complex with its cognate ligand Clr-11
To Be Published
|
|
6BL0
 
 | | Novel Modes of Inhibition of Wild-Type IDH1:Direct Covalent Modification of His315 with Cmpd11 | | Descriptor: | (5aS,6S,8S,9aS)-2-(benzenecarbonyl)-6-methyl-7-oxo-9a-phenyl-4,5,5a,6,7,8,9,9a-octahydro-2H-benzo[g]indazole-8-carbonitrile, ISOCITRIC ACID, Isocitrate dehydrogenase [NADP] cytoplasmic, ... | | Authors: | Jakob, C.G, Qiu, W. | | Deposit date: | 2017-11-09 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Novel Modes of Inhibition of Wild-Type Isocitrate Dehydrogenase 1 (IDH1): Direct Covalent Modification of His315.
J. Med. Chem., 61, 2018
|
|
6BL2
 
 | | Novel Modes of Inhibition of Wild-Type IDH1: Direct Covalent Modification of His315 with Cmpd15 | | Descriptor: | 3-[(6aS,7S,9S,10aS)-9-cyano-7-methyl-8-oxo-2-(phenylamino)-6,6a,7,8,9,10-hexahydrobenzo[h]quinazolin-10a(5H)-yl]benzoic acid, CALCIUM ION, ISOCITRIC ACID, ... | | Authors: | Jakob, C.G, Qiu, W. | | Deposit date: | 2017-11-09 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Novel Modes of Inhibition of Wild-Type Isocitrate Dehydrogenase 1 (IDH1): Direct Covalent Modification of His315.
J. Med. Chem., 61, 2018
|
|
6BKX
 
 | | Novel Modes of Inhibition of Wild-Type IDH1: Direct Covalent Modification of His315 with Cmpd1 | | Descriptor: | (6aS,7S,9R,10aS)-7,10a-dimethyl-8-oxo-2-(phenylamino)-5,6,6a,7,8,9,10,10a-octahydrobenzo[h]quinazoline-9-carbonitrile, CALCIUM ION, ISOCITRIC ACID, ... | | Authors: | Jakob, C.G, Qiu, W. | | Deposit date: | 2017-11-09 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Novel Modes of Inhibition of Wild-Type Isocitrate Dehydrogenase 1 (IDH1): Direct Covalent Modification of His315.
J. Med. Chem., 61, 2018
|
|
6BKY
 
 | | Novel Binding Modes of Inhibition of Wild-Type IDH1: Allosteric Inhibition with Cmpd2 | | Descriptor: | (2S)-2-hydroxybutanedioic acid, 4,5,6,7-TETRABROMO-1H,3H-BENZIMIDAZOL-2-ONE, Isocitrate dehydrogenase [NADP] cytoplasmic, ... | | Authors: | Jakob, C.G, Qiu, W. | | Deposit date: | 2017-11-09 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Novel Modes of Inhibition of Wild-Type Isocitrate Dehydrogenase 1 (IDH1): Direct Covalent Modification of His315.
J. Med. Chem., 61, 2018
|
|
6BL1
 
 | | Novel Modes of Inhibition of Wild-Type IDH1: Direct Covalent Modification of His315 with Cmpd13 | | Descriptor: | (6aS,7S,9S,10aS)-7-methyl-8-oxo-10a-phenyl-2-(phenylamino)-5,6,6a,7,8,9,10,10a-octahydrobenzo[h]quinazoline-9-carbonitrile, CALCIUM ION, ISOCITRIC ACID, ... | | Authors: | Jakob, C.G, Qiu, W. | | Deposit date: | 2017-11-09 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Novel Modes of Inhibition of Wild-Type Isocitrate Dehydrogenase 1 (IDH1): Direct Covalent Modification of His315.
J. Med. Chem., 61, 2018
|
|
6BKZ
 
 | | Novel Modes of Inhibition of Wild-Type IDH1: Non-equivalent Allosteric Inhibition with Cmpd3 | | Descriptor: | (7R)-1-[(4-fluorophenyl)methyl]-N-{3-[(1R)-1-hydroxyethyl]phenyl}-7-methyl-5-(1H-pyrrole-2-carbonyl)-4,5,6,7-tetrahydro-1H-pyrazolo[4,3-c]pyridine-3-carboxamide, Isocitrate dehydrogenase [NADP] cytoplasmic, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Jakob, C.G, Qiu, W. | | Deposit date: | 2017-11-09 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Novel Modes of Inhibition of Wild-Type Isocitrate Dehydrogenase 1 (IDH1): Direct Covalent Modification of His315.
J. Med. Chem., 61, 2018
|
|
7A1B
 
 | | Crystal structure of human protein kinase CK2alpha' (CSNK2A2 gene product) in complex with the ATP-competitive inhibitor 5,6-dibromo-1H-triazolo[4,5-b]pyridine | | Descriptor: | 1,2-ETHANEDIOL, 5,6-dibromo-1H-triazolo[4,5-b]pyridine, CHLORIDE ION, ... | | Authors: | Niefind, K, Lindenblatt, D, Toelzer, C, Bretner, M, Chojnacki, K, Wielechowska, M, Winska, P. | | Deposit date: | 2020-08-12 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.287 Å) | | Cite: | Synthesis, biological properties and structural study of new halogenated azolo[4,5-b]pyridines as inhibitors of CK2 kinase.
Bioorg.Chem., 106, 2021
|
|
