4J71
 
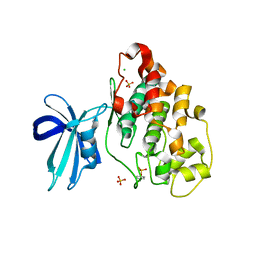 | | Crystal Structure of GSK3b in complex with inhibitor 1R | | Descriptor: | (2R)-2-methyl-1,4-dihydropyrido[2,3-b]pyrazin-3(2H)-one, CHLORIDE ION, Glycogen synthase kinase-3 beta, ... | | Authors: | Zhan, C, Wang, Y, Wach, J, Sheehan, P, Zhong, C, Harris, R, Patskovsky, Y, Bishop, J, Haggarty, S, Ramek, A, Berry, K, O'Herin, C, Koehler, A.N, Hung, A.W, Young, D.W, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-02-12 | | Release date: | 2013-03-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Fragment-based approach using diversity-oriented synthesis yields a GSK3b inhibitor
To be Published
|
|
1PG9
 
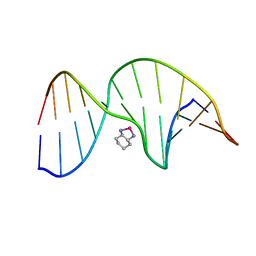 | | NMR Solution Structure of an Oxaliplatin 1,2-d(GG) Intrastrand Cross-Link in a DNA Dodecamer Duplex | | Descriptor: | 5'-D(*CP*CP*TP*CP*AP*GP*GP*CP*CP*TP*CP*C)-3', 5'-D(*GP*GP*AP*GP*GP*CP*CP*TP*GP*AP*GP*G)-3', CYCLOHEXANE-1(R),2(R)-DIAMINE-PLATINUM(II) | | Authors: | Wu, Y, Pradhan, P, Havener, J, Chaney, S.G, Campbel, S.L. | | Deposit date: | 2003-05-28 | | Release date: | 2004-07-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of an oxaliplatin 1,2-d(GG) intrastrand cross-link in a DNA dodecamer duplex
J.Mol.Biol., 341, 2004
|
|
1W74
 
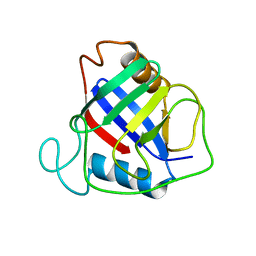 | | X-ray structure of peptidyl-prolyl cis-trans isomerase A, PpiA, Rv0009, from Mycobacterium tuberculosis. | | Descriptor: | PEPTIDYL-PROLYL CIS-TRANS ISOMERASE A | | Authors: | Henriksson, L.M, Johansson, P, Unge, T, Mowbray, S.L. | | Deposit date: | 2004-08-27 | | Release date: | 2004-10-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | X-Ray Structure of Peptidyl-Prolyl Cis-Trans Isomerase a from Mycobacterium Tuberculosis
Eur.J.Biochem., 271, 2004
|
|
2K0Q
 
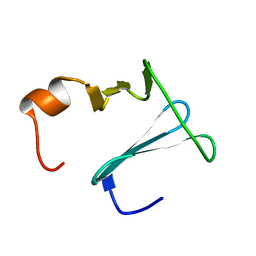 | | Solution structure of CopK, a periplasmic protein involved in copper resistance in Cupriavidus metallidurans CH34 | | Descriptor: | Putative uncharacterized protein copK | | Authors: | Bersch, B, Favier, A, Schanda, P, Coves, J, van Aelst, S, Vallaeys, T, Wattiez, R, Mergeay, M. | | Deposit date: | 2008-02-12 | | Release date: | 2008-05-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Molecular structure and metal-binding properties of the periplasmic CopK protein expressed in Cupriavidus metallidurans CH34 during copper challenge.
J.Mol.Biol., 380, 2008
|
|
4AJM
 
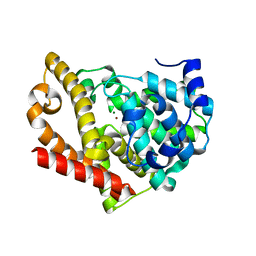 | | Development of a plate-based optical biosensor methodology to identify PDE10 fragment inhibitors | | Descriptor: | 3-AMINO-6-FLUORO-2-[4-(2-METHYLPYRIDIN-4-YL)PHENYL]-N-(METHYLSULFONYL)QUINOLINE-4-CARBOXAMIDE, CAMP AND CAMP-INHIBITED CGMP 3', 5'-CYCLIC PHOSPHODIESTERASE 10A, ... | | Authors: | Geschwindner, S, Johansson, P, Spadola, L, Akerud, T, Back, E, Hillertz, P, Horsefeld, R, Scott, C, Spear, N, Tian, G, Tigerstrom, A, Aharony, D, Albert, J.S. | | Deposit date: | 2012-02-16 | | Release date: | 2013-03-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Development of a Plate-Based Optical Biosensor Methodology to Identify Pde10 Fragment Inhibitors
To be Published
|
|
2L3Z
 
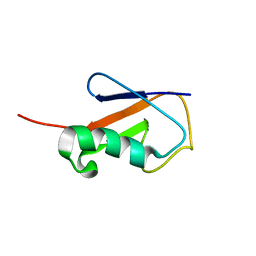 | | Proton-Detected 4D DREAM Solid-State NMR Structure of Ubiquitin | | Descriptor: | Ubiquitin | | Authors: | Huber, M, Hiller, S, Schanda, P, Ernst, M, Bockmann, A, Verel, R, Meier, B.H. | | Deposit date: | 2010-09-27 | | Release date: | 2011-02-16 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | A Proton-Detected 4D Solid-State NMR Experiment for Protein Structure Determination.
Chemphyschem, 12, 2011
|
|
2VEO
 
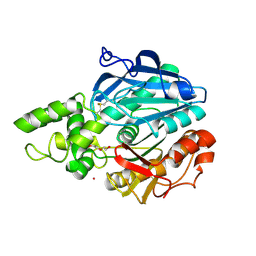 | | X-ray structure of Candida antarctica lipase A in its closed state. | | Descriptor: | GLYCEROL, LIPASE A, TETRAETHYLENE GLYCOL, ... | | Authors: | Ericsson, D.J, Kasrayan, A, Johansson, P, Bergfors, T, Sandstrom, A.G, Backvall, J.E, Mowbray, S.L. | | Deposit date: | 2007-10-25 | | Release date: | 2007-11-06 | | Last modified: | 2015-04-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-Ray Structure of Candida Antarctica Lipase a Shows a Novel Lid Structure and a Likely Mode of Interfacial Activation.
J.Mol.Biol., 376, 2008
|
|
2WTO
 
 | | Crystal Structure of Apo-form Czce from C. metallidurans CH34 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, ORF131 PROTEIN | | Authors: | Haertlein, I, Girard, E, Sarret, G, Hazemann, J, Gourhant, P, Kahn, R, Coves, J. | | Deposit date: | 2009-09-18 | | Release date: | 2010-08-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Evidence for Conformational Changes Upon Copper Binding to Cupriavidus Metallidurans Czce.
Biochemistry, 49, 2010
|
|
4OV4
 
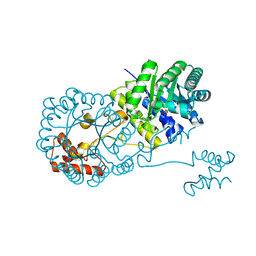 | | Isopropylmalate synthase binding with ketoisovalerate | | Descriptor: | 2-isopropylmalate synthase, 3-METHYL-2-OXOBUTANOIC ACID, ZINC ION | | Authors: | Zhang, Z, Wu, J, Wang, C, Zhang, P. | | Deposit date: | 2014-02-20 | | Release date: | 2014-08-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Subdomain II of alpha-isopropylmalate synthase is essential for activity: inferring a mechanism of feedback inhibition.
J.Biol.Chem., 289, 2014
|
|
4G91
 
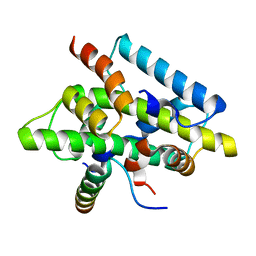 | | CCAAT-binding complex from Aspergillus nidulans | | Descriptor: | HAPB protein, HapE, Transcription factor HapC (Eurofung) | | Authors: | Huber, E.M, Scharf, D.H, Hortschansky, P, Groll, M, Brakhage, A.A. | | Deposit date: | 2012-07-23 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | DNA Minor Groove Sensing and Widening by the CCAAT-Binding Complex.
Structure, 20, 2012
|
|
4B77
 
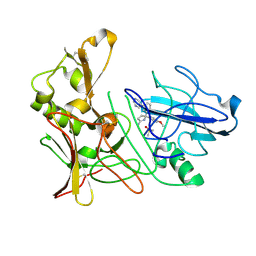 | | Aminoimidazoles as BACE-1 Inhibitors: From De Novo Design to Ab- lowering in Brain | | Descriptor: | (5R)-5-(4-methoxyphenyl)-5-(3-pyrimidin-5-ylphenyl)-3,4-dihydropyrrol-2-amine, BETA-SECRETASE 1, DIMETHYL SULFOXIDE | | Authors: | Gravenfors, Y, Blid, J, Ginman, T, Karlstrom, S, Kihlstrom, J, Kolmodin, K, Lindstrom, J, Berg, S, Kieseritzky, F, Slivo, C, Swahn, B, Viklund, J, Olsson, L, Johansson, P, Eketjall, S, Falting, J, Jeppsson, F, Stromberg, K, Janson, J, Rahm, F. | | Deposit date: | 2012-08-16 | | Release date: | 2013-06-26 | | Last modified: | 2013-07-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Core Refinement Toward Permeable Beta-Secretase (Bace-1) Inhibitors with Low Herg Activity.
J.Med.Chem., 56, 2013
|
|
4B70
 
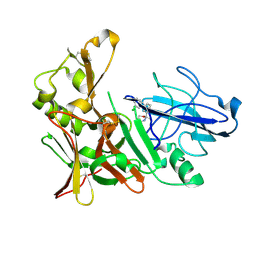 | | Aminoimidazoles as BACE-1 Inhibitors: From De Novo Design to Ab- lowering in Brain | | Descriptor: | (2S)-2-[3-(3-chlorophenyl)phenyl]-2-methyl-5,6-dihydro-1,3-oxazin-4-amine, BETA-SECRETASE 1 | | Authors: | Gravenfors, Y, Blid, J, Ginman, T, Karlstrom, S, Kihlstrom, J, Kolmodin, K, Lindstrom, J, Berg, S, Kieseritzky, F, Slivo, C, Swahn, B, Viklund, J, Olsson, L, Johansson, P, Eketjall, S, Falting, J, Jeppsson, F, Stromberg, K, Janson, J, Rahm, F. | | Deposit date: | 2012-08-16 | | Release date: | 2013-06-26 | | Last modified: | 2013-07-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Core Refinement Toward Permeable Beta-Secretase (Bace-1) Inhibitors with Low Herg Activity.
J.Med.Chem., 56, 2013
|
|
2WTP
 
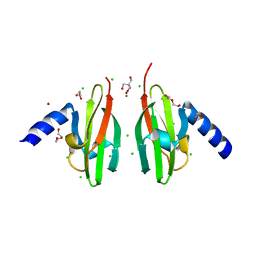 | | Crystal Structure of Cu-form Czce from C. metallidurans CH34 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Haertlein, I, Girard, E, Sarret, G, Hazemann, J, Gourhant, P, Kahn, R, Coves, J. | | Deposit date: | 2009-09-18 | | Release date: | 2010-08-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Evidence for Conformational Changes Upon Copper Binding to Cupriavidus Metallidurans Czce.
Biochemistry, 49, 2010
|
|
4G92
 
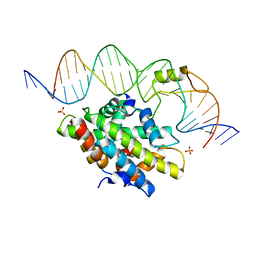 | | CCAAT-binding complex from Aspergillus nidulans with DNA | | Descriptor: | DNA, HAPB protein, HapE, ... | | Authors: | Huber, E.M, Scharf, D.H, Hortschansky, P, Groll, M, Brakhage, A.A. | | Deposit date: | 2012-07-23 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | DNA Minor Groove Sensing and Widening by the CCAAT-Binding Complex.
Structure, 20, 2012
|
|
2WA2
 
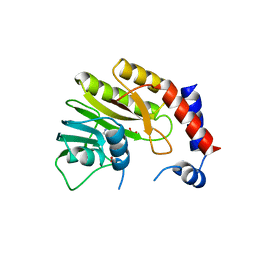 | | Structure of the methyltransferase domain from Modoc Virus, a Flavivirus with No Known Vector (NKV) | | Descriptor: | NON-STRUCTURAL PROTEIN 5, S-ADENOSYLMETHIONINE, SULFATE ION | | Authors: | Jansson, A.M, Johansson, P, Jones, T.A. | | Deposit date: | 2009-02-02 | | Release date: | 2009-08-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the Methyltransferase Domain from the Modoc Virus, a Flavivirus with No Known Vector.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
4B78
 
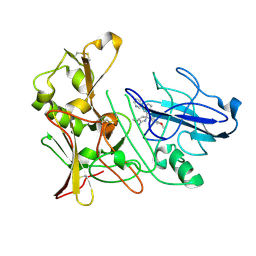 | | Aminoimidazoles as BACE-1 Inhibitors: From De Novo Design to Ab- lowering in Brain | | Descriptor: | (3R,5R)-3-methoxy-5-(4-methoxyphenyl)-5-(3-pyridin-3-ylphenyl)-3,4-dihydropyrrol-2-amine, BETA-SECRETASE 1 | | Authors: | Gravenfors, Y, Blid, J, Ginman, T, Karlstrom, S, Kihlstrom, J, Kolmodin, K, Lindstrom, J, Berg, S, Kieseritzky, F, Slivo, C, Swahn, B, Viklund, J, Olsson, L, Johansson, P, Eketjall, S, Falting, J, Jeppsson, F, Stromberg, K, Janson, J, Rahm, F. | | Deposit date: | 2012-08-16 | | Release date: | 2013-06-26 | | Last modified: | 2013-07-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Core Refinement Toward Permeable Beta-Secretase (Bace-1) Inhibitors with Low Herg Activity.
J.Med.Chem., 56, 2013
|
|
3S32
 
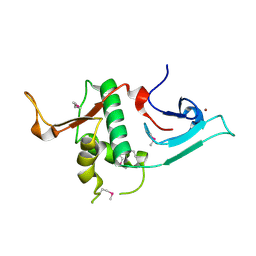 | | Crystal structure of Ash2L N-terminal domain | | Descriptor: | Set1/Ash2 histone methyltransferase complex subunit ASH2, ZINC ION | | Authors: | Sarvan, S, Avdic, V, Tremblay, V, Chaturvedi, C.-P, Zhang, P, Lanouette, S, Blais, A, Brunzelle, J.S, Brand, M, Couture, J.-F. | | Deposit date: | 2011-05-17 | | Release date: | 2011-06-08 | | Last modified: | 2012-01-11 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of the trithorax group protein ASH2L reveals a forkhead-like DNA binding domain.
Nat.Struct.Mol.Biol., 18, 2011
|
|
4GH8
 
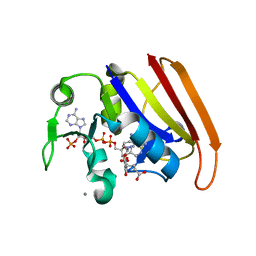 | | Crystal structure of a 'humanized' E. coli dihydrofolate reductase | | Descriptor: | CALCIUM ION, Dihydrofolate reductase, METHOTREXATE, ... | | Authors: | French, J.B, Liu, C.T, Hanoian, P, Pringle, T.H, Hammes-Schiffer, S, Benkovic, S.J. | | Deposit date: | 2012-08-07 | | Release date: | 2013-06-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Functional significance of evolving protein sequence in dihydrofolate reductase from bacteria to humans.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4OV9
 
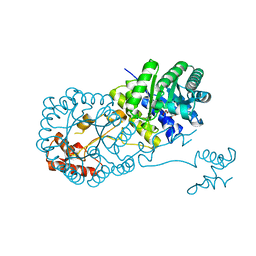 | | Structure of isopropylmalate synthase binding with alpha-isopropylmalate | | Descriptor: | (2S)-2-hydroxy-2-(propan-2-yl)butanedioic acid, ZINC ION, isopropylmalate synthase | | Authors: | Zhang, Z, Wu, J, Wang, C, Zhang, P. | | Deposit date: | 2014-02-20 | | Release date: | 2014-08-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Subdomain II of alpha-isopropylmalate synthase is essential for activity: inferring a mechanism of feedback inhibition.
J.Biol.Chem., 289, 2014
|
|
4B72
 
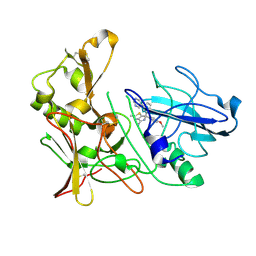 | | Aminoimidazoles as BACE-1 Inhibitors: From De Novo Design to Ab- lowering in Brain | | Descriptor: | (6R)-6-(4-methoxyphenyl)-2-methyl-6-(3-pyrimidin-5-ylphenyl)pyrrolo[3,4-d][1,3]thiazol-4-amine, BETA-SECRETASE 1 | | Authors: | Gravenfors, Y, Blid, J, Ginman, T, Karlstrom, S, Kihlstrom, J, Kolmodin, K, Lindstrom, J, Berg, S, Kieseritzky, F, Slivo, C, Swahn, B, Viklund, J, Olsson, L, Johansson, P, Eketjall, S, Falting, J, Jeppsson, F, Stromberg, K, Janson, J, Rahm, F. | | Deposit date: | 2012-08-16 | | Release date: | 2013-06-26 | | Last modified: | 2013-07-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Core Refinement Toward Permeable Beta-Secretase (Bace-1) Inhibitors with Low Herg Activity.
J.Med.Chem., 56, 2013
|
|
2X98
 
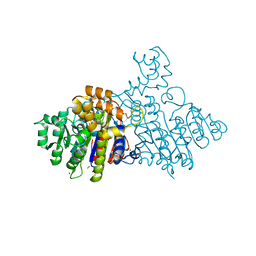 | | H.SALINARUM ALKALINE PHOSPHATASE | | Descriptor: | ALKALINE PHOSPHATASE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Wende, A, Johansson, P, Grininger, M, Oesterhelt, D. | | Deposit date: | 2010-03-14 | | Release date: | 2010-05-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Biochemical Characterization of a Halophilic Archaeal Alkaline Phosphatase.
J.Mol.Biol., 400, 2010
|
|
4RFS
 
 | | Structure of a pantothenate energy coupling factor transporter | | Descriptor: | Energy-coupling factor transporter ATP-binding protein EcfA1, Energy-coupling factor transporter ATP-binding protein EcfA2, Energy-coupling factor transporter transmembrane protein EcfT, ... | | Authors: | Zhang, M, Bao, Z, Zhao, Q, Guo, H, Xu, K, Zhang, P. | | Deposit date: | 2014-09-27 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.232 Å) | | Cite: | Structure of a pantothenate transporter and implications for ECF module sharing and energy coupling of group II ECF transporters.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4AL1
 
 | | Crystal structure of Human PS-1 GSH-analog complex | | Descriptor: | L-gamma-glutamyl-S-(2-biphenyl-4-yl-2-oxoethyl)-L-cysteinylglycine, PALMITIC ACID, PROSTAGLANDIN E SYNTHASE, ... | | Authors: | Sjogren, T, Nord, J, Ek, M, Johansson, P, Liu, G, Geschwindner, S. | | Deposit date: | 2012-02-29 | | Release date: | 2013-02-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Microsomal Prostaglandin E2 Synthase Provides Insight Into Diversity in the Mapeg Superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1QCO
 
 | | CRYSTAL STRUCTURE OF FUMARYLACETOACETATE HYDROLASE COMPLEXED WITH FUMARATE AND ACETOACETATE | | Descriptor: | ACETOACETIC ACID, CALCIUM ION, FUMARIC ACID, ... | | Authors: | Timm, D.E, Mueller, H.A, Bhanumoorthy, P, Harp, J.M, Bunick, G.J. | | Deposit date: | 1999-05-17 | | Release date: | 2000-06-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and mechanism of a carbon-carbon bond hydrolase.
Structure Fold.Des., 7, 1999
|
|
4B1D
 
 | | New Aminoimidazoles as BACE-1 Inhibitors: From Rational Design to Ab- lowering in Brain | | Descriptor: | (2S)-2-(4-methoxy-3,5-dimethylphenyl)-5-methyl-2-(3-pyrimidin-5-ylphenyl)-2H-imidazol-4-amine, ACETATE ION, BETA-SECRETASE 1 | | Authors: | Rahm, F, Blid, J, Ginman, T, Karlstrom, S, Kihlstrom, J, Kolmodin, K, Lindstrom, J, von Berg, S, von Kieseritzky, F, Slivo, C, Swahn, B, Viklund, J, Olsson, L, Johansson, P, Eketjall, S, Falting, J, Jeppsson, F, Stromberg, K, Janson, J, Gravenfors, Y. | | Deposit date: | 2012-07-10 | | Release date: | 2012-10-10 | | Last modified: | 2018-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | New aminoimidazoles as beta-secretase (BACE-1) inhibitors showing amyloid-beta (A beta ) lowering in brain.
J. Med. Chem., 55, 2012
|
|
