2OBO
 
 | | Structure of HEPATITIS C VIRAL NS3 protease domain complexed with NS4A peptide and ketoamide SCH476776 | | Descriptor: | BETA-MERCAPTOETHANOL, HCV NS3 protease, HCV NS4A peptide, ... | | Authors: | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers Jr, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R, Arasappan, A, Bennett, F, Bogen, S.F, Chen, K, Jao, E, Liu, Y, Love, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | Deposit date: | 2006-12-19 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
2OC8
 
 | | Structure of Hepatitis C Viral NS3 protease domain complexed with NS4A peptide and ketoamide SCH503034 | | Descriptor: | BETA-MERCAPTOETHANOL, Hepatitis C virus, ZINC ION, ... | | Authors: | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R.S, Arasappan, A, Bennett, F, Bogen, S.L, Chen, K, Jao, E, Liu, Y.T, Lovey, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | Deposit date: | 2006-12-20 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
6O2Q
 
 | | Acetylated Microtubules | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Eshun-Wilson, L, Zhang, R, Portran, D, Nachury, M.V, Toso, D, Lohr, T, Vendruscolo, M, Bonomi, M, Fraser, J.S, Nogales, E. | | Deposit date: | 2019-02-24 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Effects of alpha-tubulin acetylation on microtubule structure and stability.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7SOM
 
 | | Ciliary C2 central pair apparatus isolated from Chlamydomonas reinhardtii | | Descriptor: | Cilia- and flagella-associated protein 20, FAP147, FAP178, ... | | Authors: | Gui, M, Wang, X, Dutcher, S.K, Brown, A, Zhang, R. | | Deposit date: | 2021-11-01 | | Release date: | 2022-04-13 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Ciliary central apparatus structure reveals mechanisms of microtubule patterning.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7SQC
 
 | | Ciliary C1 central pair apparatus isolated from Chlamydomonas reinhardtii | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CPC1, Calmodulin, ... | | Authors: | Gui, M, Wang, X, Dutcher, S.K, Brown, A, Zhang, R. | | Deposit date: | 2021-11-05 | | Release date: | 2022-04-13 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Ciliary central apparatus structure reveals mechanisms of microtubule patterning.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6O2R
 
 | | Deacetylated Microtubules | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Eshun-Wilson, L, Zhang, R, Portran, D, Nachury, M.V, Toso, D, Lohr, T, Vendruscolo, M, Bonomi, M, Fraser, J.S, Nogales, E. | | Deposit date: | 2019-02-24 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Effects of alpha-tubulin acetylation on microtubule structure and stability.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
2G5C
 
 | | Crystal Structure of Prephenate Dehydrogenase from Aquifex aeolicus | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, prephenate dehydrogenase | | Authors: | Sun, W, Singh, S, Zhang, R, Turnbull, J.L, Christendat, D. | | Deposit date: | 2006-02-22 | | Release date: | 2006-03-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Prephenate Dehydrogenase from Aquifex aeolicus: Insights into the Catalytic Mechanism
J.Biol.Chem., 281, 2006
|
|
2OQW
 
 | | The crystal structure of sortase B from B.anthracis in complex with AAEK1 | | Descriptor: | Sortase B | | Authors: | Wu, R, Zhang, R, Marresso, A.W, Schneewind, O, Joachimiak, A. | | Deposit date: | 2007-02-01 | | Release date: | 2007-06-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Activation of inhibitors by sortase triggers irreversible modification of the active site.
J.Biol.Chem., 282, 2007
|
|
2OQZ
 
 | | The crystal structure of sortase B from B.anthracis in complex with AAEK2 | | Descriptor: | ACETIC ACID, Sortase B | | Authors: | Wu, R, Zhang, R, Maresso, A.W, Schneewind, O, Joachimiak, A. | | Deposit date: | 2007-02-01 | | Release date: | 2007-06-19 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Activation of inhibitors by sortase triggers irreversible modification of the active site.
J.Biol.Chem., 282, 2007
|
|
2PPW
 
 | |
3CQV
 
 | | Crystal structure of Reverb beta in complex with heme | | Descriptor: | Nuclear receptor subfamily 1 group D member 2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Xu, X, Dong, A, Pardee, K.I, Reinking, J, Krause, H, Schuetz, A, Zhang, R, Cui, H, Edwards, A, Arrowsmith, C.H, Weigelt, J, Bountra, C, Savchenko, A, Botchkarev, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-04-03 | | Release date: | 2008-08-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis of gas-responsive transcription by the human nuclear hormone receptor REV-ERBbeta.
Plos Biol., 7, 2009
|
|
3D6K
 
 | | The crystal structure of a putative aminotransferase from Corynebacterium diphtheriae | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Putative aminotransferase, ... | | Authors: | Tan, K, Zhang, R, Duggan, E, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-19 | | Release date: | 2008-07-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of a putative aminotransferase from Corynebacterium diphtheriae
To be Published
|
|
6JKM
 
 | | Crystal structure of BubR1 kinase domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Lin, L, Ye, S, Huang, Y, Liu, X, Zhang, R, Yao, X. | | Deposit date: | 2019-03-01 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | BubR1 phosphorylates CENP-E as a switch enabling the transition from lateral association to end-on capture of spindle microtubules.
Cell Res., 29, 2019
|
|
6JKK
 
 | | Crystal structure of BubR1 kinase domain | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Mitotic checkpoint control protein kinase BUB1 | | Authors: | Lin, L, Ye, S, Huang, Y, Liu, X, Zhang, R, Yao, X. | | Deposit date: | 2019-03-01 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | BubR1 phosphorylates CENP-E as a switch enabling the transition from lateral association to end-on capture of spindle microtubules.
Cell Res., 29, 2019
|
|
8Q20
 
 | | Crystal structure of Vanadium-dependent haloperoxidase R425D mutant (A. marina) | | Descriptor: | PHOSPHATE ION, SULFATE ION, Vanadium-dependent bromoperoxidase, ... | | Authors: | Zeides, P, Bellmannn-Sickert, K, Zhang, R, Seel, C.J, Most, V, Schroeder, C.T, Groll, M, Gulder, T. | | Deposit date: | 2023-08-01 | | Release date: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Unraveling the Molecular Basis of Substrate Specificity and Halogen Activation in Vanadium-Dependent Haloperoxidases
to be published
|
|
8Q21
 
 | | Crystal structure of Vanadium-dependent haloperoxidase R425S mutant (A. marina) | | Descriptor: | PHOSPHATE ION, SODIUM ION, Vanadium-dependent bromoperoxidase, ... | | Authors: | Zeides, P, Bellmannn-Sickert, K, Zhang, R, Seel, C.J, Most, V, Schroeder, C.T, Groll, M, Gulder, T. | | Deposit date: | 2023-08-01 | | Release date: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Unraveling the Molecular Basis of Substrate Specificity and Halogen Activation in Vanadium-Dependent Haloperoxidases
to be published
|
|
8Q22
 
 | | Crystal structure of Vanadium-dependent haloperoxidase R425S mutant in complex with 1,3,5-trimethoxybenzene (A. marina) | | Descriptor: | 1,3,5-trimethoxybenzene, PHOSPHATE ION, SODIUM ION, ... | | Authors: | Zeides, P, Bellmannn-Sickert, K, Zhang, R, Seel, C.J, Most, V, Schroeder, C.T, Groll, M, Gulder, T. | | Deposit date: | 2023-08-01 | | Release date: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Unraveling the Molecular Basis of Substrate Specificity and Halogen Activation in Vanadium-Dependent Haloperoxidases
to be published
|
|
2I5U
 
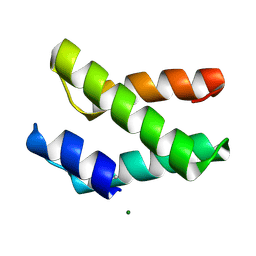 | | Crystal structure of DnaD domain protein from Enterococcus faecalis. Structural genomics target APC85179 | | Descriptor: | DnaD domain protein, MAGNESIUM ION | | Authors: | Wu, R, Zhang, R, Bargassa, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-08-25 | | Release date: | 2006-11-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.5 A crystal structure of a DnaD domain protein from Enterococcus Faecalis
To be Published, 2006
|
|
1D9K
 
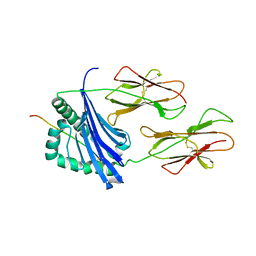 | | CRYSTAL STRUCTURE OF COMPLEX BETWEEN D10 TCR AND PMHC I-AK/CA | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CONALBUMIN PEPTIDE, ... | | Authors: | Reinherz, E.L, Tan, K, Tang, L, Kern, P, Liu, J.-H, Xiong, Y, Hussey, R.E, Smolyar, A, Hare, B, Zhang, R, Joachimiak, A, Chang, H.-C, Wagner, G, Wang, J.-H. | | Deposit date: | 1999-10-28 | | Release date: | 1999-12-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The crystal structure of a T cell receptor in complex with peptide and MHC class II.
Science, 286, 1999
|
|
4DX8
 
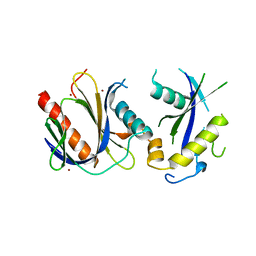 | | ICAP1 in complex with KRIT1 N-terminus | | Descriptor: | BROMIDE ION, Integrin beta-1-binding protein 1, Krev interaction trapped protein 1 | | Authors: | Liu, W, Draheim, K, Zhang, R, Calderwood, D.A, Boggon, T.J. | | Deposit date: | 2012-02-27 | | Release date: | 2013-01-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Mechanism for KRIT1 Release of ICAP1-Mediated Suppression of Integrin Activation.
Mol.Cell, 49, 2013
|
|
4DXA
 
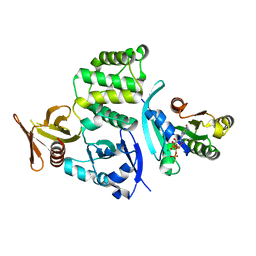 | | Co-crystal structure of Rap1 in complex with KRIT1 | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Krev interaction trapped protein 1, MAGNESIUM ION, ... | | Authors: | Li, X, Zhang, R, Boggon, T.J. | | Deposit date: | 2012-02-27 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Basis for Small G Protein Effector Interaction of Ras-related Protein 1 (Rap1) and Adaptor Protein Krev Interaction Trapped 1 (KRIT1).
J.Biol.Chem., 287, 2012
|
|
4DX9
 
 | | ICAP1 in complex with integrin beta 1 cytoplasmic tail | | Descriptor: | Integrin beta-1, Integrin beta-1-binding protein 1 | | Authors: | Liu, W, Draheim, K, Zhang, R, Calderwood, D.A, Boggon, T.J. | | Deposit date: | 2012-02-27 | | Release date: | 2013-01-09 | | Last modified: | 2020-09-02 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Mechanism for KRIT1 Release of ICAP1-Mediated Suppression of Integrin Activation.
Mol.Cell, 49, 2013
|
|
4F7G
 
 | |
1EG3
 
 | | STRUCTURE OF A DYSTROPHIN WW DOMAIN FRAGMENT IN COMPLEX WITH A BETA-DYSTROGLYCAN PEPTIDE | | Descriptor: | DYSTROPHIN | | Authors: | Huang, X, Poy, F, Zhang, R, Joachimiak, A, Sudol, M, Eck, M.J. | | Deposit date: | 2000-02-11 | | Release date: | 2000-08-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a WW domain containing fragment of dystrophin in complex with beta-dystroglycan.
Nat.Struct.Biol., 7, 2000
|
|
6U42
 
 | | Natively decorated ciliary doublet microtubule | | Descriptor: | DC1, DC2, DC3, ... | | Authors: | Ma, M, Stoyanova, M, Rademacher, G, Dutcher, S.K, Brown, A, Zhang, R. | | Deposit date: | 2019-08-22 | | Release date: | 2019-11-13 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the Decorated Ciliary Doublet Microtubule.
Cell, 179, 2019
|
|
