7X7R
 
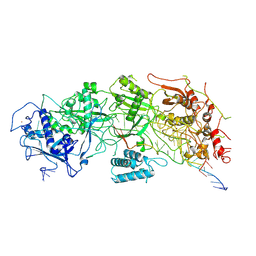 | | Cryo-EM structure of a bacterial protein | | Descriptor: | RAMP superfamily protein, RNA (36-MER), RNA (5'-R(P*AP*GP*UP*CP*CP*GP*GP*GP*GP*CP*AP*GP*AP*AP*AP*AP*UP*UP*GP*G)-3'), ... | | Authors: | Yu, G, Wang, X, Deng, Z, Zhang, H. | | Deposit date: | 2022-03-10 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure and function of a bacterial type III-E CRISPR-Cas7-11 complex.
Nat Microbiol, 7, 2022
|
|
5L0A
 
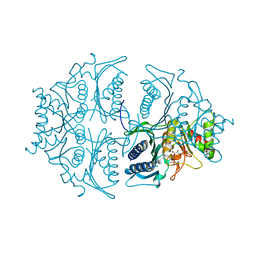 | | Human muscle fructose-1,6-bisphosphatase E69Q mutant in active R-state in complex with fructose-1,6-bisphosphate | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase isozyme 2 | | Authors: | Barciszewski, J, Wisniewski, J, Kolodziejczyk, R, Dzugaj, A, Jaskolski, M, Rakus, D. | | Deposit date: | 2016-07-27 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Structural studies of human muscle FBPase
To Be Published
|
|
7VR1
 
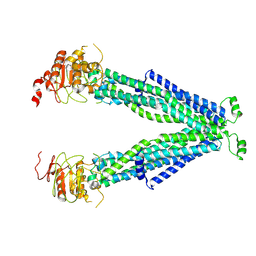 | |
2WZK
 
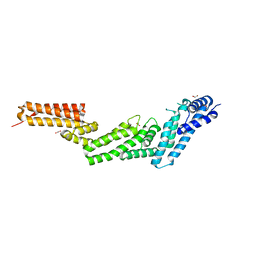 | | Structure of the Cul5 N-terminal domain at 2.05A resolution. | | Descriptor: | 1,2-ETHANEDIOL, CULLIN-5 | | Authors: | Muniz, J.R.C, Ayinampudi, V, Zhang, Y, Babon, J.J, Chaikuad, A, Krojer, T, Pike, A.C.W, Vollmar, M, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Bullock, A.N. | | Deposit date: | 2009-11-30 | | Release date: | 2009-12-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Molecular Architecture of the Ankyrin Socs Box Family of Cul5-Dependent E3 Ubiquitin Ligases
J.Mol.Biol., 425, 2013
|
|
7D8M
 
 | | Crystal structure of DyP | | Descriptor: | Dye-decolorizing peroxidase, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | He, C, Jia, R, Wang, T, Li, L.Q. | | Deposit date: | 2020-10-08 | | Release date: | 2021-08-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Revealing two important tryptophan residues with completely different roles in a dye-decolorizing peroxidase from Irpex lacteus F17.
Biotechnol Biofuels, 14, 2021
|
|
1GMH
 
 | |
7V2S
 
 | | Crystal structure of juvenile hormone acid methyltransferase JHAMT isoform3 from silkworm | | Descriptor: | Methyltranfer_dom domain-containing protein | | Authors: | Guo, P.C, Zhang, Y.S, Zhang, l, Xu, H.Y. | | Deposit date: | 2021-08-09 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.133 Å) | | Cite: | Structural characterization and functional analysis of juvenile hormone acid methyltransferase JHAMT3 from the silkworm, Bombyx mori.
Insect Biochem.Mol.Biol., 151, 2022
|
|
1GCD
 
 | |
3C2T
 
 | | Evolution of chlorella virus dUTPase | | Descriptor: | DEOXYURIDINE-5'-DIPHOSPHATE, Deoxyuridine triphosphatase, MAGNESIUM ION | | Authors: | Yamanishi, M, Homma, K, Zhang, Y, Etten, L.V.J, Moriyama, H. | | Deposit date: | 2008-01-25 | | Release date: | 2009-02-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystallization and crystal-packing studies of Chlorella virus deoxyuridine triphosphatase.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
5K55
 
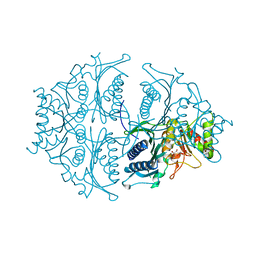 | | Human muscle fructose-1,6-bisphosphatase E69Q mutant in active R-state in complex with fructose-6-phosphate | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase isozyme 2 | | Authors: | Barciszewski, J, Wisniewski, J, Kolodziejczyk, R, Dzugaj, A, Jaskolski, M, Rakus, D. | | Deposit date: | 2016-05-23 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.977 Å) | | Cite: | Structural studies of human muscle FBPase
To Be Published
|
|
7W1Y
 
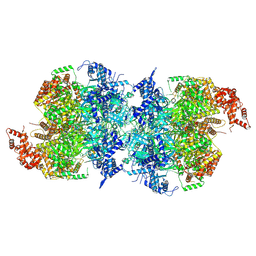 | | Human MCM double hexamer bound to natural DNA duplex (polyAT/polyTA) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, DNA (49-MER), ... | | Authors: | Li, J, Dong, J, Dang, S, Zhai, Y. | | Deposit date: | 2021-11-21 | | Release date: | 2023-02-08 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | The human pre-replication complex is an open complex.
Cell, 186, 2023
|
|
7WKP
 
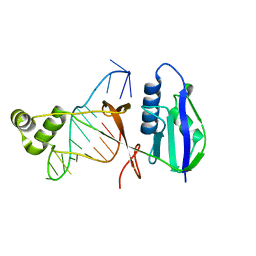 | | ICP1 Csy4 | | Descriptor: | 3' stem loop crRNA, Csy4 | | Authors: | Zhang, M, Peng, R. | | Deposit date: | 2022-01-10 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanistic insights into DNA binding and cleavage by a compact type I-F CRISPR-Cas system in bacteriophage.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7WKO
 
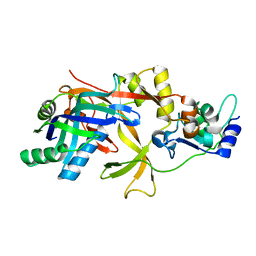 | | ICP1 Csy1-2 complex | | Descriptor: | Csy1, Csy2 | | Authors: | Zhang, M, Peng, R. | | Deposit date: | 2022-01-10 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanistic insights into DNA binding and cleavage by a compact type I-F CRISPR-Cas system in bacteriophage.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7XP1
 
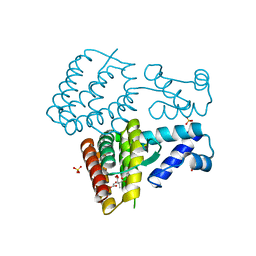 | | Crystal structure of PmiR from Pseudomonas aeruginosa | | Descriptor: | ALPHA-METHYLISOCITRIC ACID, GLYCEROL, Probable transcriptional regulator, ... | | Authors: | Zhang, Y.X, Liang, H.H, Gan, J.H. | | Deposit date: | 2022-05-02 | | Release date: | 2023-04-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | PmiR senses 2-methylisocitrate levels to regulate bacterial virulence in Pseudomonas aeruginosa.
Sci Adv, 8, 2022
|
|
7XP0
 
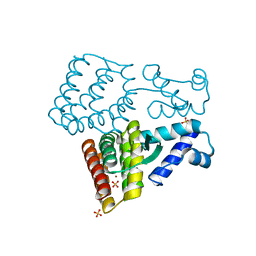 | |
7EEB
 
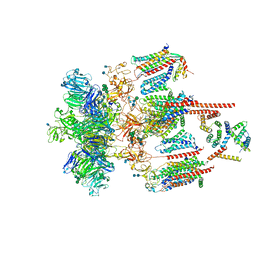 | | Structure of the CatSpermasome | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wu, J.P, Ke, M. | | Deposit date: | 2021-03-18 | | Release date: | 2021-07-28 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of a mammalian sperm cation channel complex.
Nature, 595, 2021
|
|
7EBG
 
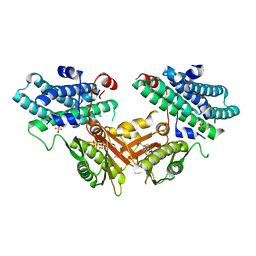 | | Crystal structure of human pyruvate dehydrogenase kinase 4 in complex with compound 7 | | Descriptor: | 3,3-dimethyl-7-(methylamino)-1H-indol-2-one, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Orita, T, Doi, S, Iwanaga, T, Adachi, T. | | Deposit date: | 2021-03-09 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Fragment-based lead discovery to identify novel inhibitors that target the ATP binding site of pyruvate dehydrogenase kinases.
Bioorg.Med.Chem., 44, 2021
|
|
7EAT
 
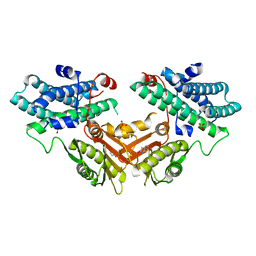 | | Crystal structure of human pyruvate dehydrogenase kinase 4 in complex with compound 1 | | Descriptor: | 1,3-dihydro-2H-indol-2-one, SULFATE ION, [Pyruvate dehydrogenase (acetyl-transferring)] kinase isozyme 4, ... | | Authors: | Orita, T, Doi, S, Iwanaga, T, Adachi, T. | | Deposit date: | 2021-03-08 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Fragment-based lead discovery to identify novel inhibitors that target the ATP binding site of pyruvate dehydrogenase kinases.
Bioorg.Med.Chem., 44, 2021
|
|
7EBB
 
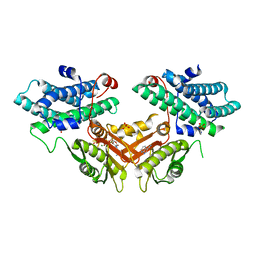 | | Crystal structure of human pyruvate dehydrogenase kinase 4 in complex with compound 2 | | Descriptor: | 1H-pyrrolo[2,3-b]pyridine-3-carbonitrile, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Orita, T, Doi, S, Iwanaga, T, Adachi, T. | | Deposit date: | 2021-03-09 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fragment-based lead discovery to identify novel inhibitors that target the ATP binding site of pyruvate dehydrogenase kinases.
Bioorg.Med.Chem., 44, 2021
|
|
7EA0
 
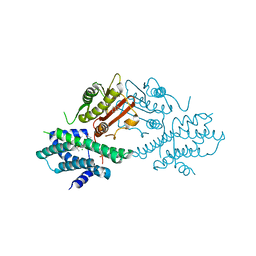 | | Crystal structure of human pyruvate dehydrogenase kinase 2 in complex with compound 1 | | Descriptor: | 1,3-dihydro-2H-indol-2-one, ACETATE ION, CHLORIDE ION, ... | | Authors: | Orita, T, Doi, S, Iwanaga, T, Adachi, T. | | Deposit date: | 2021-03-05 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Fragment-based lead discovery to identify novel inhibitors that target the ATP binding site of pyruvate dehydrogenase kinases.
Bioorg.Med.Chem., 44, 2021
|
|
7EAS
 
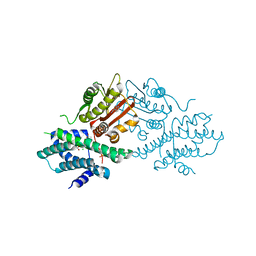 | | Crystal structure of human pyruvate dehydrogenase kinase 2 in complex with compound 2 | | Descriptor: | 1H-pyrrolo[2,3-b]pyridine-3-carbonitrile, ACETATE ION, CHLORIDE ION, ... | | Authors: | Orita, T, Doi, S, Iwanaga, T, Adachi, T. | | Deposit date: | 2021-03-08 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Fragment-based lead discovery to identify novel inhibitors that target the ATP binding site of pyruvate dehydrogenase kinases.
Bioorg.Med.Chem., 44, 2021
|
|
7EBH
 
 | | Crystal structure of human pyruvate dehydrogenase kinase 2 in complex with compound 13 | | Descriptor: | 5-bromanyl-2-methyl-6-propyl-7H-pyrrolo[2,3-d]pyrimidine, ACETATE ION, CHLORIDE ION, ... | | Authors: | Orita, T, Doi, S, Iwanaga, T, Adachi, T. | | Deposit date: | 2021-03-09 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Fragment-based lead discovery to identify novel inhibitors that target the ATP binding site of pyruvate dehydrogenase kinases.
Bioorg.Med.Chem., 44, 2021
|
|
7Y75
 
 | | SIT1-ACE2-BA.2 RBD | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shen, Y.P, Li, Y.N, Zhang, Y.Y, Yan, R.H. | | Deposit date: | 2022-06-21 | | Release date: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of ACE2-SIT1 recognized by Omicron variants of SARS-CoV-2.
Cell Discov, 8, 2022
|
|
7Y76
 
 | | SIT1-ACE2-BA.5 RBD | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shen, Y.P, Li, Y.N, Zhang, Y.Y, Yan, R.H. | | Deposit date: | 2022-06-21 | | Release date: | 2023-01-04 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of ACE2-SIT1 recognized by Omicron variants of SARS-CoV-2.
Cell Discov, 8, 2022
|
|
4J1U
 
 | | Crystal structure of antibody 93F3 unstable variant | | Descriptor: | antibody 93F3 Heavy chain, antibody 93F3 Light chain | | Authors: | Wang, F. | | Deposit date: | 2013-02-02 | | Release date: | 2013-03-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Somatic hypermutation maintains antibody thermodynamic stability during affinity maturation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
