1NIB
 
 | | THE STRUCTURE OF CU-NITRITE REDUCTASE FROM ACHROMOBACTER CYCLOCLASTES AT FIVE PH VALUES, WITH NITRITE BOUND AND WITH TYPE II CU DEPLETED | | Descriptor: | COPPER (II) ION, NITRITE REDUCTASE | | Authors: | Adman, E.T, Godden, J.W, Turley, S. | | Deposit date: | 1995-07-03 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of copper-nitrite reductase from Achromobacter cycloclastes at five pH values, with NO2- bound and with type II copper depleted.
J.Biol.Chem., 270, 1995
|
|
6VBN
 
 | | Crystal Structure of hTDO2 bound to inhibitor GNE1 | | Descriptor: | 1,5-anhydro-2,3-dideoxy-3-[(5S)-5H-imidazo[5,1-a]isoindol-5-yl]-D-threo-pentitol, PROTOPORPHYRIN IX CONTAINING FE, Tryptophan 2,3-dioxygenase | | Authors: | Harris, S.F, Oh, A. | | Deposit date: | 2019-12-19 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Implementation of the CYP Index for the Design of Selective Tryptophan-2,3-dioxygenase Inhibitors.
Acs Med.Chem.Lett., 11, 2020
|
|
1NID
 
 | | THE STRUCTURE OF CU-NITRITE REDUCTASE FROM ACHROMOBACTER CYCLOCLASTES AT FIVE PH VALUES, WITH NITRITE BOUND AND WITH TYPE II CU DEPLETED | | Descriptor: | COPPER (II) ION, NITRITE ION, NITRITE REDUCTASE | | Authors: | Adman, E.T, Godden, J.W, Turley, S. | | Deposit date: | 1995-07-03 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of copper-nitrite reductase from Achromobacter cycloclastes at five pH values, with NO2- bound and with type II copper depleted.
J.Biol.Chem., 270, 1995
|
|
1NIA
 
 | | THE STRUCTURE OF CU-NITRITE REDUCTASE FROM ACHROMOBACTER CYCLOCLASTES AT FIVE PH VALUES, WITH NITRITE BOUND AND WITH TYPE II CU DEPLETED | | Descriptor: | COPPER (II) ION, NITRITE REDUCTASE | | Authors: | Adman, E.T, Godden, J.W, Turley, S. | | Deposit date: | 1995-07-03 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of copper-nitrite reductase from Achromobacter cycloclastes at five pH values, with NO2- bound and with type II copper depleted.
J.Biol.Chem., 270, 1995
|
|
1NIC
 
 | | THE STRUCTURE OF CU-NITRITE REDUCTASE FROM ACHROMOBACTER CYCLOCLASTES AT FIVE PH VALUES, WITH NITRITE BOUND AND WITH TYPE II CU DEPLETED | | Descriptor: | COPPER (II) ION, NITRITE REDUCTASE, SULFATE ION | | Authors: | Adman, E.T, Godden, J.W, Turley, S. | | Deposit date: | 1995-07-03 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of copper-nitrite reductase from Achromobacter cycloclastes at five pH values, with NO2- bound and with type II copper depleted.
J.Biol.Chem., 270, 1995
|
|
6NC2
 
 | | AMC011 v4.2 SOSIP Env trimer in complex with fusion peptide targeting antibody ACS202 fragment antigen binding | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, AMC011 v4.2 SOSIP gp120, ... | | Authors: | Cottrell, C.A, Ozorowski, G, Yuan, M, Copps, J, Wilson, I.A, Ward, A.B. | | Deposit date: | 2018-12-10 | | Release date: | 2019-06-19 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Conformational Plasticity in the HIV-1 Fusion Peptide Facilitates Recognition by Broadly Neutralizing Antibodies.
Cell Host Microbe, 25, 2019
|
|
4CTS
 
 | |
6NCP
 
 | | Crystal structure of HIV-1 broadly neutralizing antibody ACS202 | | Descriptor: | ACS202 Fab heavy chain, ACS202 Fab light chain, GLYCINE, ... | | Authors: | Yuan, M, Wilson, I.A. | | Deposit date: | 2018-12-11 | | Release date: | 2019-06-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Conformational Plasticity in the HIV-1 Fusion Peptide Facilitates Recognition by Broadly Neutralizing Antibodies.
Cell Host Microbe, 25, 2019
|
|
6NC3
 
 | | AMC011 v4.2 SOSIP Env trimer in complex with fusion peptide targeting antibody VRC34 fragment antigen binding | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 Env AMC011 v4.2 SOSIP gp120, ... | | Authors: | Cottrell, C.A, Ozorowski, G, Torres, J.L, Ward, A.B. | | Deposit date: | 2018-12-10 | | Release date: | 2019-06-19 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Conformational Plasticity in the HIV-1 Fusion Peptide Facilitates Recognition by Broadly Neutralizing Antibodies.
Cell Host Microbe, 25, 2019
|
|
1NIE
 
 | | THE STRUCTURE OF CU-NITRITE REDUCTASE FROM ACHROMOBACTER CYCLOCLASTES AT FIVE PH VALUES, WITH NITRITE BOUND AND WITH TYPE II CU DEPLETED | | Descriptor: | COPPER (II) ION, NITRITE REDUCTASE | | Authors: | Adman, E.T, Godden, J.W, Turley, S. | | Deposit date: | 1995-07-03 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of copper-nitrite reductase from Achromobacter cycloclastes at five pH values, with NO2- bound and with type II copper depleted.
J.Biol.Chem., 270, 1995
|
|
4CSC
 
 | |
1CSC
 
 | |
1BMK
 
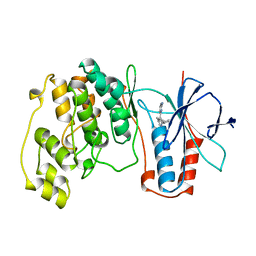 | | THE COMPLEX STRUCTURE OF THE MAP KINASE P38/SB218655 | | Descriptor: | 4-(FLUOROPHENYL)-1-CYCLOPROPYLMETHYL-5-(2-AMINO-4-PYRIMIDINYL)IMIDAZOLE, PROTEIN (MAP KINASE P38) | | Authors: | Wang, Z, Canagarajah, B, Boehm, J.C, Kassis, S, Cobb, M.H, Young, P.R, Abdel-Meguid, S, Adams, J.L, Goldsmith, E.J. | | Deposit date: | 1998-07-23 | | Release date: | 1999-07-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of inhibitor selectivity in MAP kinases.
Structure, 6, 1998
|
|
1BL6
 
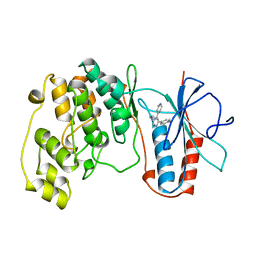 | | THE COMPLEX STRUCTURE OF THE MAP KINASE P38/SB216995 | | Descriptor: | 4-(4-FLUOROPHENYL)-1-CYCLOROPROPYLMETHYL-5-(4-PYRIDYL)-IMIDAZOLE, PROTEIN (MAP KINASE P38) | | Authors: | Wang, Z, Canagarajah, B.J, Boehm, J.C, Kassis, S, Cobb, M.H, Young, P.R, Abdel-Meguid, S, Adams, J.L, Goldsmith, E.J. | | Deposit date: | 1998-07-11 | | Release date: | 1999-07-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of inhibitor selectivity in MAP kinases.
Structure, 6, 1998
|
|
1BL7
 
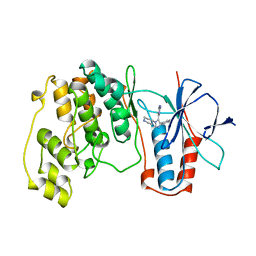 | | THE COMPLEX STRUCTURE OF THE MAP KINASE P38/SB220025 | | Descriptor: | 4-(4-FLUOROPHENYL)-1-(4-PIPERIDINYL)-5-(2-AMINO-4-PYRIMIDINYL)-IMIDAZOLE, PROTEIN (MAP KINASE P38) | | Authors: | Wang, Z, Canagarajah, B.J, Boehm, J.C, Kassis, S, Cobb, M.H, Young, P.R, Abdel-Meguid, S, Adams, J.L, Goldsmith, E.J. | | Deposit date: | 1998-07-23 | | Release date: | 1999-07-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of inhibitor selectivity in MAP kinases.
Structure, 6, 1998
|
|
1A9U
 
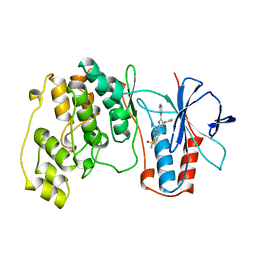 | | THE COMPLEX STRUCTURE OF THE MAP KINASE P38/SB203580 | | Descriptor: | 4-[5-(4-FLUORO-PHENYL)-2-(4-METHANESULFINYL-PHENYL)-3H-IMIDAZOL-4-YL]-PYRIDINE, MAP KINASE P38 | | Authors: | Wang, Z, Canagarajah, B, Boehm, J.C, Kassis, S, Cobb, M.H, Young, P.R, Abdel-Meguid, S, Adams, J.L, Goldsmith, E.J. | | Deposit date: | 1998-04-10 | | Release date: | 1999-04-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of inhibitor selectivity in MAP kinases.
Structure, 6, 1998
|
|
1CSI
 
 | |
2I9G
 
 | | DNA Polymerase Beta with a Benzo[c]phenanthrene diol epoxide adducted guanine base | | Descriptor: | (1S)-1,2,3,4-TETRAHYDRO-BENZO[C]PHENANTHRENE-2,3,4-TRIOL, 5'-D(*CP*CP*GP*AP*CP*GP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3', ... | | Authors: | Batra, V.K, Wilson, S.H, Beard, W.A, Pedersen, L.C. | | Deposit date: | 2006-09-05 | | Release date: | 2006-10-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of DNA polymerase beta with a benzo[c]phenanthrene diol epoxide-adducted template exhibits mutagenic features.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1CSH
 
 | |
4Z9S
 
 | |
6CTS
 
 | |
3VTS
 
 | |
3R1A
 
 | | Closed crystal structure of cytochrome P450 2B4 covalently bound to the mechanism-based inactivator tert-butylphenylacetylene | | Descriptor: | (4-tert-butylphenyl)acetaldehyde, Cytochrome P450 2B4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Gay, S.C, Zhang, H, Stout, C.D, Hollenberg, P.F, Halpert, J.R. | | Deposit date: | 2011-03-09 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural Analysis of Mammalian Cytochrome P450 2B4 Covalently Bound to the Mechanism-Based Inactivator tert-Butylphenylacetylene: Insight into Partial Enzymatic Activity.
Biochemistry, 50, 2011
|
|
3R1B
 
 | | Open crystal structure of cytochrome P450 2B4 covalently bound to the mechanism-based inactivator tert-butylphenylacetylene | | Descriptor: | (4-tert-butylphenyl)acetaldehyde, 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, Cytochrome P450 2B4, ... | | Authors: | Gay, S.C, Zhang, H, Stout, C.D, Hollenberg, P.F, Halpert, J.R. | | Deposit date: | 2011-03-09 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Analysis of Mammalian Cytochrome P450 2B4 Covalently Bound to the Mechanism-Based Inactivator tert-Butylphenylacetylene: Insight into Partial Enzymatic Activity.
Biochemistry, 50, 2011
|
|
4FOP
 
 | | Crystal Structure of Peptidyl-tRNA hydrolase from Acinetobacter baumannii at 1.86 A resolution | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Kaushik, S, Kumar, S, Singh, N, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-06-21 | | Release date: | 2012-07-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The Mode of Inhibitor Binding to Peptidyl-tRNA Hydrolase: Binding Studies and Structure Determination of Unbound and Bound Peptidyl-tRNA Hydrolase from Acinetobacter baumannii
Plos One, 8, 2013
|
|
