7L0G
 
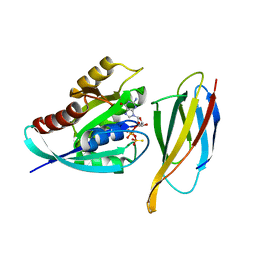 | | Monobody 12VC1 Bound to HRAS(G12C) | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Teng, K.W, Hattori, T, Tsai, S, Koide, S. | | Deposit date: | 2020-12-11 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Selective and noncovalent targeting of RAS mutants for inhibition and degradation.
Nat Commun, 12, 2021
|
|
6JW2
 
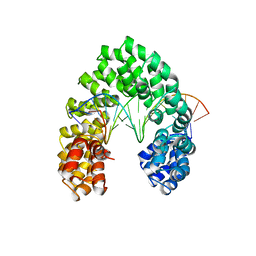 | | Universal RVD R* accommodates 5hmC via water-mediated interactions | | Descriptor: | DNA (5'-D(*AP*GP*AP*GP*AP*CP*GP*CP*GP*AP*AP*GP*GP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*CP*CP*CP*TP*TP*(5HC)P*GP*CP*GP*TP*CP*TP*CP*T)-3'), TAL effector | | Authors: | Liu, L, Yi, C. | | Deposit date: | 2019-04-18 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Structural Insights into the Specific Recognition of 5-methylcytosine and 5-hydroxymethylcytosine by TAL Effectors.
J.Mol.Biol., 432, 2020
|
|
6JVZ
 
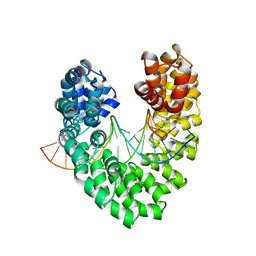 | | RVD HA specifically contacts 5mC through van der Waals interactions | | Descriptor: | DNA (5'-D(*AP*GP*AP*GP*AP*CP*GP*CP*GP*AP*AP*GP*GP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*CP*CP*CP*TP*TP*(5CM)P*GP*CP*GP*TP*CP*TP*CP*T)-3'), TAL effector | | Authors: | Liu, L, Yi, C. | | Deposit date: | 2019-04-18 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural Insights into the Specific Recognition of 5-methylcytosine and 5-hydroxymethylcytosine by TAL Effectors.
J.Mol.Biol., 432, 2020
|
|
8H3Z
 
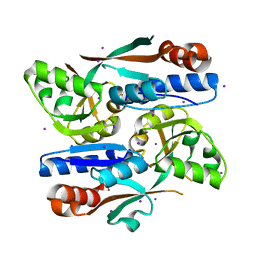 | |
8JIY
 
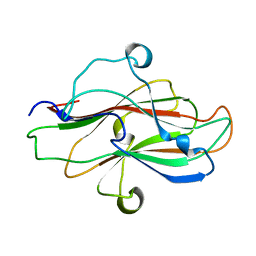 | | A carbohydrate binding domain of a putative chondroitinase | | Descriptor: | DUF4955 domain-containing protein | | Authors: | Liu, G.C, Chang, Y.G. | | Deposit date: | 2023-05-29 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Identification and structural characterization of a novel chondroitin sulfate-specific carbohydrate-binding module: The first member of a new family, CBM100.
Int.J.Biol.Macromol., 255, 2024
|
|
8JEU
 
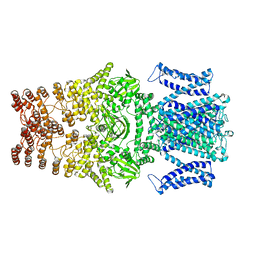 | |
8JEC
 
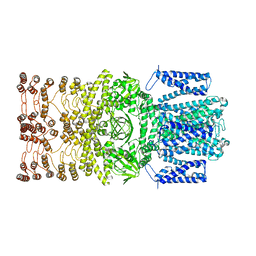 | |
8JET
 
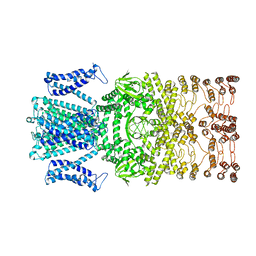 | |
8IZL
 
 | |
8W6V
 
 | |
7W6L
 
 | | The crystal structure of MLL3-RBBP5-ASH2L in complex with H3K4me0 peptide | | Descriptor: | Histone H3.3C, Histone-lysine N-methyltransferase 2C, Retinoblastoma-binding protein 5, ... | | Authors: | Zhao, L, Li, Y, Chen, Y. | | Deposit date: | 2021-12-01 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural basis for product specificities of MLL family methyltransferases.
Mol.Cell, 82, 2022
|
|
7W6A
 
 | | Crystal structure of the MLL1 (N3861I/Q3867L/C3882SS)-RBBP5-ASH2L complex | | Descriptor: | Histone-lysine N-methyltransferase 2A, Retinoblastoma-binding protein 5, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Zhao, L, Li, Y, Chen, Y. | | Deposit date: | 2021-12-01 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural basis for product specificities of MLL family methyltransferases.
Mol.Cell, 82, 2022
|
|
7W6I
 
 | | The crystal structure of MLL1 (N3861I/Q3867L/C3882SS)-RBBP5-ASH2L in complex with H3K4me1 peptide | | Descriptor: | Histone H3.3C, Histone-lysine N-methyltransferase 2A, Retinoblastoma-binding protein 5, ... | | Authors: | Zhao, L, Li, Y, Chen, Y. | | Deposit date: | 2021-12-01 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structural basis for product specificities of MLL family methyltransferases.
Mol.Cell, 82, 2022
|
|
7W67
 
 | | The crystal structure of MLL1 (N3861I/Q3867L/C3882SS)-RBBP5-ASH2L in complex with H3K4me0 peptide | | Descriptor: | Histone H3.3C, Histone-lysine N-methyltransferase 2A, Retinoblastoma-binding protein 5, ... | | Authors: | Zhao, L, Li, Y, Chen, Y. | | Deposit date: | 2021-12-01 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | Structural basis for product specificities of MLL family methyltransferases.
Mol.Cell, 82, 2022
|
|
7W6J
 
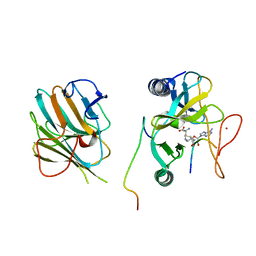 | | The crystal structure of MLL1 (N3861I/Q3867L/C3882SS)-RBBP5-ASH2L in complex with H3K4me2 peptide | | Descriptor: | Histone H3.3C, Histone-lysine N-methyltransferase 2A, Retinoblastoma-binding protein 5, ... | | Authors: | Zhao, L, Li, Y, Chen, Y. | | Deposit date: | 2021-12-01 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structural basis for product specificities of MLL family methyltransferases.
Mol.Cell, 82, 2022
|
|
8VH5
 
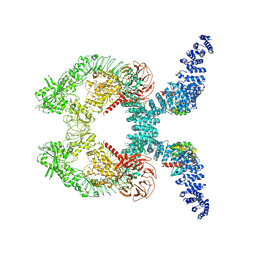 | | Cryo-EM structure of Rab12-LRRK2 complex in the LRRK2 dimer state | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Leucine-rich repeat serine/threonine-protein kinase 2, MAGNESIUM ION, ... | | Authors: | Zhu, H, Sun, J. | | Deposit date: | 2023-12-30 | | Release date: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | RAB12-LRRK2 complex suppresses primary ciliogenesis and regulates centrosome homeostasis in astrocytes.
Nat Commun, 15, 2024
|
|
8VH4
 
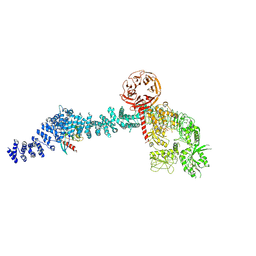 | | Cryo-EM structure of Rab12-LRRK2 complex in the LRRK2 monomer state | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Leucine-rich repeat serine/threonine-protein kinase 2, MAGNESIUM ION, ... | | Authors: | Zhu, H, Sun, J. | | Deposit date: | 2023-12-30 | | Release date: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | RAB12-LRRK2 complex suppresses primary ciliogenesis and regulates centrosome homeostasis in astrocytes.
Nat Commun, 15, 2024
|
|
7DK8
 
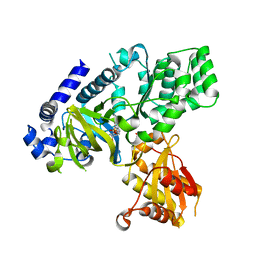 | | Crystal structure of OsGH3-8 with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Probable indole-3-acetic acid-amido synthetase GH3.8 | | Authors: | Zhang, Y.K, Xu, G.L, Ming, Z.H. | | Deposit date: | 2020-11-23 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal structure of the acyl acid amido synthetase GH3-8 from Oryza sativa.
Biochem.Biophys.Res.Commun., 534, 2021
|
|
7D5I
 
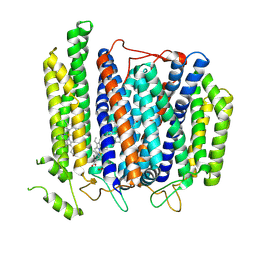 | | Structure of Mycobacterium smegmatis bd complex in the apo-form. | | Descriptor: | CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, Cytochrome D ubiquinol oxidase subunit 1, HEME B/C, ... | | Authors: | Wang, W, Gong, H, Gao, Y, Zhou, X, Rao, Z. | | Deposit date: | 2020-09-26 | | Release date: | 2021-06-23 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Cryo-EM structure of mycobacterial cytochrome bd reveals two oxygen access channels.
Nat Commun, 12, 2021
|
|
3VJE
 
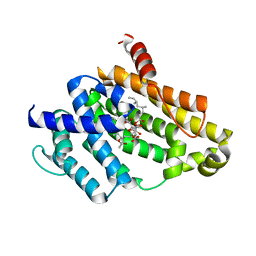 | | Crystal structure of the Y248A mutant of C(30) carotenoid dehydrosqualene synthase from Staphylococcus aureus in complex with zaragozic acid A | | Descriptor: | Dehydrosqualene synthase, Zaragozic acid A | | Authors: | Liu, C.I, Jeng, W.Y, Chang, W.J, Wang, A.H.J. | | Deposit date: | 2011-10-14 | | Release date: | 2012-04-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Binding modes of zaragozic acid A to human squalene synthase and staphylococcal dehydrosqualene synthase
J.Biol.Chem., 287, 2012
|
|
7DAJ
 
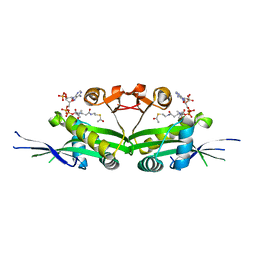 | | The crystal structure of serotonin N-acetyltransferase in complex with acetyl-CoA from Oryza Sativa | | Descriptor: | ACETYL COENZYME *A, Serotonin N-acetyltransferase 1, chloroplastic | | Authors: | Zhou, Y.Z, Liao, L.J, Tang, T, Guo, Y, Liu, X.K, Liu, B, Zhao, Y.C. | | Deposit date: | 2020-10-16 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Molecular Dynamics Analysis of Plant Serotonin N-Acetyltransferase Reveal an Acid/Base-Assisted Catalysis in Melatonin Biosynthesis.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7DAI
 
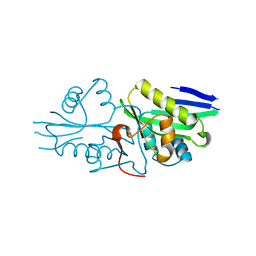 | | The crystal structure of a serotonin N-acetyltransferase from Oryza Sativa | | Descriptor: | Serotonin N-acetyltransferase 1, chloroplastic | | Authors: | Zhou, Y.Z, Liao, L.J, Tang, T, Guo, Y, Liu, X.K, Liu, B, Zhao, Y.C. | | Deposit date: | 2020-10-16 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Molecular Dynamics Analysis of Plant Serotonin N-Acetyltransferase Reveal an Acid/Base-Assisted Catalysis in Melatonin Biosynthesis.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7DAL
 
 | | The crystal structure of a serotonin N-acetyltransferase in complex with serotonin and acetyl-CoA from Oryza Sativa | | Descriptor: | ACETYL COENZYME *A, SEROTONIN, Serotonin N-acetyltransferase 1, ... | | Authors: | Zhou, Y.Z, Liao, L.J, Tang, T, Guo, Y, Liu, X.K, Liu, B, Zhao, Y.C. | | Deposit date: | 2020-10-16 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Molecular Dynamics Analysis of Plant Serotonin N-Acetyltransferase Reveal an Acid/Base-Assisted Catalysis in Melatonin Biosynthesis.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7DAK
 
 | | The crystal structure of a serotonin N-acetyltransferase in complex with 5-Methoxytryptamine and acetyl-CoA from Oryza Sativa | | Descriptor: | 2-(5-methoxy-1H-indol-3-yl)ethanamine, ACETYL COENZYME *A, Serotonin N-acetyltransferase 1, ... | | Authors: | Zhou, Y.Z, Liao, L.J, Tang, T, Guo, Y, Liu, X.K, Liu, B, Zhao, Y.C. | | Deposit date: | 2020-10-16 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and Molecular Dynamics Analysis of Plant Serotonin N-Acetyltransferase Reveal an Acid/Base-Assisted Catalysis in Melatonin Biosynthesis.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
8W71
 
 | | Structural basis of chorismate isomerization by Arabidopsis isochorismate synthase ICS1 | | Descriptor: | (3R,4R)-3-[(1-carboxyethenyl)oxy]-4-hydroxycyclohexa-1,5-diene-1-carboxylic acid, FORMIC ACID, Isochorismate synthase 1, ... | | Authors: | Su, Z.H, Ming, Z.H. | | Deposit date: | 2023-08-30 | | Release date: | 2024-05-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.121 Å) | | Cite: | Structural basis of chorismate isomerization by Arabidopsis ISOCHORISMATE SYNTHASE1.
Plant Physiol., 196, 2024
|
|
