6VF4
 
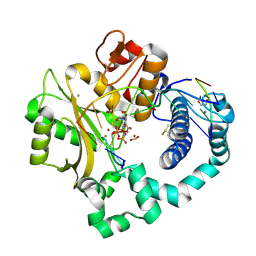 | | DNA Polymerase Mu, 8-oxorGTP:At Reaction State Ternary Complex, 50 mM Mn2+ (30 min) | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2020-01-03 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | DNA Polymerase Mu, 8-oxorGTP:At Reaction State Ternary Complex, 50 mM Mn2+ (30 min)
To be published
|
|
6VEZ
 
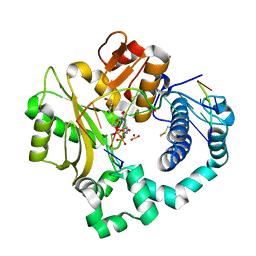 | | DNA Polymerase Mu, 8-oxorGTP:At Pre-Catalytic Ternary Complex, 20 mM Ca2+ (60 min) | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2020-01-03 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.875 Å) | | Cite: | DNA Polymerase Mu, 8-oxorGTP:At Pre-Catalytic Ternary Complex, 20 mM Ca2+ (60 min)
To be published
|
|
6VF6
 
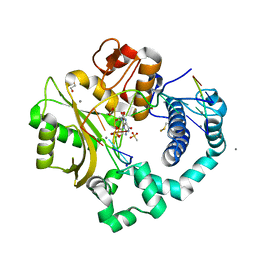 | | DNA Polymerase Mu, 8-oxorGTP:At Product State Ternary Complex, 50 mM Mn2+ (960 min) | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2020-01-03 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | DNA Polymerase Mu, 8-oxorGTP:At Product State Ternary Complex, 50 mM Mn2+ (960 min)
To be published
|
|
6VF5
 
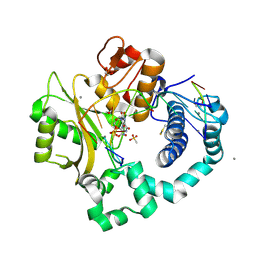 | | DNA Polymerase Mu, 8-oxorGTP:At Product State Ternary Complex, 50 mM Mn2+ (120 min) | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2020-01-03 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | DNA Polymerase Mu, 8-oxorGTP:At Product State Ternary Complex, 50 mM Mn2+ (120 min)
To be published
|
|
6VF9
 
 | | DNA Polymerase Mu, 8-oxorGTP:Ct Ternary Complex, 50 mM Mg2+ (2160 min) | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2020-01-03 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | DNA Polymerase Mu, 8-oxorGTP:Ct Ternary Complex, 50 mM Mg2+ (2160 min)
To be published
|
|
6VF7
 
 | | DNA Polymerase Mu, 8-oxodGTP:At Ground State Ternary Complex, 50 mM Mn2+ (15 min) | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2020-01-03 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.871 Å) | | Cite: | DNA Polymerase Mu, 8-oxodGTP:At Ground State Ternary Complex, 50 mM Mn2+ (15 min)
To be published
|
|
6VFC
 
 | | DNA Polymerase Mu, 8-oxorGTP:Ct Product State Ternary Complex, 50 mM Mn2+ (2160 min) | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2020-01-03 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | DNA Polymerase Mu, 8-oxorGTP:Ct Product State Ternary Complex, 50 mM Mn2+ (2160 min)
To be published
|
|
2V36
 
 | | Crystal structure of gamma-glutamyl transferase from Bacillus subtilis | | Descriptor: | GAMMA-GLUTAMYLTRANSPEPTIDASE LARGE CHAIN, GAMMA-GLUTAMYLTRANSPEPTIDASE SMALL CHAIN | | Authors: | Sharath, B, Prabhune, A.A, Suresh, C.G, Wilkinson, A.J, Brannigan, J.A. | | Deposit date: | 2007-06-13 | | Release date: | 2008-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Gamma-Glutamyl Transferase
To be Published
|
|
6VS9
 
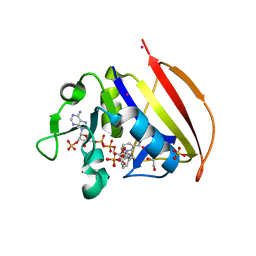 | |
5D5L
 
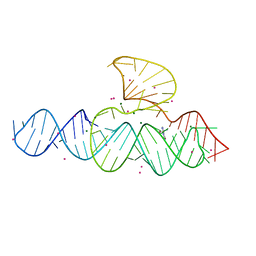 | |
5DBQ
 
 | | Crystal structure of insect thioredoxin at 1.95 Angstroms | | Descriptor: | Thioredoxin | | Authors: | Klinke, S, Tejedor, M.D, Cerutti, M.L, Giacometti, R, Otero, L.H, Goldbaum, F.A, Zavala, J.A, Wolosiuk, R.A, Pagano, E.A. | | Deposit date: | 2015-08-21 | | Release date: | 2016-08-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of insect thioredoxin at 1.95 Angstroms
To Be Published
|
|
2XO6
 
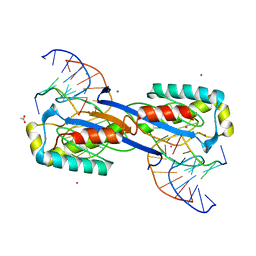 | | DEINOCOCCUS RADIODURANS ISDRA2 TRANSPOSASE Y132F MUTANT COMPLEXED WITH LEFT END RECOGNITION AND CLEAVAGE SITE | | Descriptor: | 5'-D(*TP*TP*GP*AP*TP*G)-3', ACETATE ION, CADMIUM ION, ... | | Authors: | Hickman, A.B, James, J.A, Barabas, O, Pasternak, C, Ton-Hoang, B, Chandler, M, Sommer, S, Dyda, F. | | Deposit date: | 2010-08-09 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | DNA Recognition and the Precleavage State During Single-Stranded DNA Transposition in D. Radiodurans.
Embo J., 29, 2010
|
|
6TT5
 
 | | Crystal structure of DCLRE1C/Artemis | | Descriptor: | 1,2-ETHANEDIOL, NICKEL (II) ION, Protein artemis, ... | | Authors: | Yosaatmadja, Y, Goubin, S, Newman, J.A, Mukhopadhyay, S.M.M, Dannerfjord, A.A, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2019-12-23 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and mechanistic insights into the Artemis endonuclease and strategies for its inhibition.
Nucleic Acids Res., 49, 2021
|
|
5DI7
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain in complex with an methyl-substituted A-CD ring estrogen derivative (1S,3aR,5S,7aS)-5-(4-hydroxy-2-methylphenyl)-7a-methyloctahydro-1H-inden-1-ol | | Descriptor: | (1S,3aR,5S,7aS)-5-(4-hydroxy-2-methylphenyl)-7a-methyloctahydro-1H-inden-1-ol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-08-31 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.241 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
6TD4
 
 | | IRF4 DNA-binding domain surface entropy mutant apo structure | | Descriptor: | CHLORIDE ION, Interferon regulatory factor 4 | | Authors: | Tucker, J.A, Martin, M.P, Wang, L.Z, Jennings, C, Heath, R. | | Deposit date: | 2019-11-07 | | Release date: | 2020-11-18 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Cancer-associated mutations in the IRF4 DNA-binding domain confer no disadvantage in DNA-binding affinity and may increase transcriptional activity
To Be Published
|
|
5DIG
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain in complex with a trifluoromethyl-substituted A-CD ring estrogen derivative (1S,3aR,5S,7aS)-5-[4-hydroxy-2-(trifluoromethyl)phenyl]-7a-methyloctahydro-1H-inden-1-ol | | Descriptor: | (1S,3aR,5S,7aS)-5-[4-hydroxy-2-(trifluoromethyl)phenyl]-7a-methyloctahydro-1H-inden-1-ol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-09-01 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
6T8Y
 
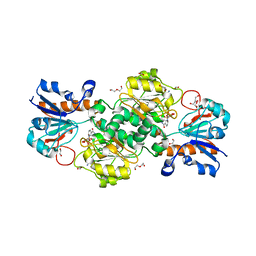 | | NAD+-dependent fungal formate dehydrogenase from Chaetomium thermophilum: A complex with the reduced form of the cofactor NADH and the substrate formate at a secondary site. | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Isupov, M.N, Yelmazer, B, De Rose, S.A, Littlechild, J.A. | | Deposit date: | 2019-10-25 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Structural insights into the NAD + -dependent formate dehydrogenase mechanism revealed from the NADH complex and the formate NAD + ternary complex of the Chaetomium thermophilum enzyme.
J.Struct.Biol., 212, 2020
|
|
5CYB
 
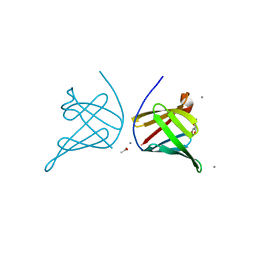 | | Structure of a lipocalin lipoprotein affecting virulence in Streptococcus pneumoniae | | Descriptor: | ACETATE ION, CALCIUM ION, GLYCEROL, ... | | Authors: | Carrasco-Lopez, C, Abdullah, M.R, Hammerschmidt, S, Hermoso, J.A. | | Deposit date: | 2015-07-30 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Unraveling the function and structure of a lipocalin lipoprotein affecting virulence in Streptococcus pneumoniae
To Be Published
|
|
5D0J
 
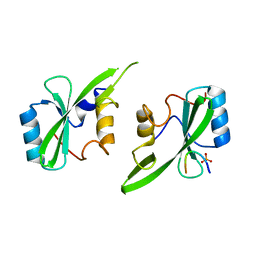 | | Grb7 SH2 with inhibitor peptide | | Descriptor: | G7-TEdFP peptide, Growth factor receptor-bound protein 7, PHOSPHATE ION | | Authors: | Gunzburg, M.J, Watson, G.M, Wilce, J.A, Wilce, M.C.J. | | Deposit date: | 2015-08-03 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Unexpected involvement of staple leads to redesign of selective bicyclic peptide inhibitor of Grb7.
Sci Rep, 6, 2016
|
|
2WCH
 
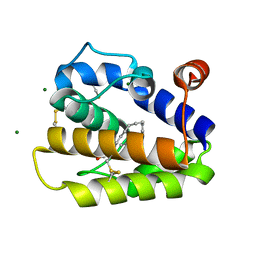 | | Structure of BMori GOBP2 (General Odorant Binding Protein 2) with bombykal | | Descriptor: | (10E,12Z)-hexadeca-10,12-dienal, GENERAL ODORANT-BINDING PROTEIN 1, MAGNESIUM ION | | Authors: | Robertson, G, Zhou, J.-J, He, X, Pickett, J.A, Field, L.M, Keep, N.H. | | Deposit date: | 2009-03-12 | | Release date: | 2009-08-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Characterisation of Bombyx Mori Odorant-Binding Proteins Reveals that a General Odorant-Binding Protein Discriminates between Sex Pheromone Components.
J.Mol.Biol., 389, 2009
|
|
2W35
 
 | | Structures of endonuclease V with DNA reveal initiation of deaminated adenine repair | | Descriptor: | 5'-D(*AP*GP*CP*CP*GP*TP)-3', 5'-D(*AP*TP*GP*CP*GP*AP*CP*IP*GP)-3', Endonuclease V, ... | | Authors: | Dalhus, B, Arvai, A.S, Rosnes, I, Olsen, O.E, Backe, P.H, Alseth, I, Gao, H, Cao, W, Tainer, J.A, Bjoras, M. | | Deposit date: | 2008-11-06 | | Release date: | 2009-01-20 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structures of Endonuclease V with DNA Reveal Initiation of Deaminated Adenine Repair.
Nat.Struct.Mol.Biol., 16, 2009
|
|
2W2D
 
 | | Crystal Structure of a Catalytically Active, Non-toxic Endopeptidase Derivative of Clostridium botulinum Toxin A | | Descriptor: | ACETATE ION, BOTULINUM NEUROTOXIN A HEAVY CHAIN, BOTULINUM NEUROTOXIN A LIGHT CHAIN, ... | | Authors: | Masuyer, G, Thiyagarajan, N, James, P.L, Marks, P.M.H, Chaddock, J.A, Acharya, K.R. | | Deposit date: | 2008-10-29 | | Release date: | 2009-03-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal Structure of a Catalytically Active, Non-Toxic Endopeptidase Derivative of Clostridium Botulinum Toxin A.
Biochem.Biophys.Res.Commun., 381, 2009
|
|
2VWT
 
 | | Crystal structure of YfaU, a metal ion dependent class II aldolase from Escherichia coli K12 - Mg-pyruvate product complex | | Descriptor: | GLYCEROL, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Rea, D, Rakus, J.F, Gerlt, J.A, Fulop, V, Bugg, T.D.H, Roper, D.I. | | Deposit date: | 2008-06-26 | | Release date: | 2008-09-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal Structure and Functional Assignment of Yfau, a Metal Ion Dependent Class II Aldolase from Escherichia Coli K12.
Biochemistry, 47, 2008
|
|
5AG5
 
 | | CRYSTAL STRUCTURE OF LEISHMANIA MAJOR N-MYRISTOYLTRANSFERASE (NMT) WITH BOUND MYRISTOYL-COA AND A THIAZOLIDINONE LIGAND | | Descriptor: | (2R)-3-benzyl-2-(1H-indazol-5-yl)-1,3-thiazolidin-4-one, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, TETRADECANOYL-COA | | Authors: | Robinson, D.A, Spinks, D, Smith, V.C, Thompson, S, Smith, A, Torrie, L.S, McElroy, S.P, Brand, S, Brenk, R, Frearson, J.A, Read, K.D, Wyatt, P.G, Gilbert, I.H. | | Deposit date: | 2015-01-29 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Development of Small-Molecule Trypanosoma Brucei N-Myristoyltransferase Inhibitors: Discovery and Optimisation of a Novel Binding Mode.
Chemmedchem, 10, 2015
|
|
2W22
 
 | | Activation Mechanism of Bacterial Thermoalkalophilic Lipases | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(2-{2-[2-(2-{2-[2-(2-{2-[4-(1,1,3,3-TETRAMETHYL-BUTYL)-PHENOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOX Y}-ETHOXY)-ETHANOL, CALCIUM ION, ... | | Authors: | Carrasco-Lopez, C, Godoy, C, De Las Rivas, B, Fernandez-Lorente, G, Palomo, J.M, Guisan, J.M, Fernandez-Lafuente, R, Hermoso, J.A. | | Deposit date: | 2008-10-23 | | Release date: | 2008-12-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Activation of bacterial thermoalkalophilic lipases is spurred by dramatic structural rearrangements.
J. Biol. Chem., 284, 2009
|
|
