2C1E
 
 | | Crystal structures of caspase-3 in complex with aza-peptide Michael acceptor inhibitors. | | Descriptor: | AZA-PEPTIDE INHIBITOR (5S, 8R, 11S)-8-(2-CARBOXYETHYL)-5-(CARBOXYMETHYL)-14-(4-ETHOXY-4-OXOBUTANOYL)-11-(1-METHYLETHYL)-3,6,9,12-TETRAOXO-1-PHENYL-2-OXA-4,7,10,13,14-PENTAAZAHEXADECAN -16-OIC ACID, ... | | Authors: | Grutter, M.G. | | Deposit date: | 2005-09-14 | | Release date: | 2006-09-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Design, Synthesis, and Evaluation of Aza-Peptide Michael Acceptors as Selective and Potent Inhibitors of Caspases-2, -3, -6, -7, -8, -9, and - 10.
J.Med.Chem., 49, 2006
|
|
6U6U
 
 | | IL36R extracellular domain in complex with BI655130 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BI00655130 Fab heavy chain, ... | | Authors: | Larson, E.T, Farrow, N.A. | | Deposit date: | 2019-08-30 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | X-ray crystal structure localizes the mechanism of inhibition of an IL-36R antagonist monoclonal antibody to interaction with Ig1 and Ig2 extra cellular domains.
Protein Sci., 29, 2020
|
|
2CCM
 
 | | X-ray structure of Calexcitin from Loligo pealeii at 1.8A | | Descriptor: | CALCIUM ION, CALEXCITIN | | Authors: | Erskine, P.T, Beaven, G.D.E, Wood, S.P, Fox, G, Vernon, J, Giese, K.P, Cooper, J.B. | | Deposit date: | 2006-01-16 | | Release date: | 2006-01-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the Neuronal Protein Calexcitin Suggests a Mode of Interaction in Signalling Pathways of Learning and Memory.
J.Mol.Biol., 357, 2006
|
|
5WDO
 
 | | H-Ras bound to GMP-PNP at 277K | | Descriptor: | CALCIUM ION, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Cofsky, J.C, Bandaru, P, Gee, C.L, Kuriyan, J. | | Deposit date: | 2017-07-05 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Deconstruction of the Ras switching cycle through saturation mutagenesis.
Elife, 6, 2017
|
|
5WDQ
 
 | | H-Ras mutant L120A bound to GMP-PNP at 100K | | Descriptor: | ACETATE ION, CALCIUM ION, GTPase HRas, ... | | Authors: | Bandaru, P, Gee, C.L, Kuriyan, J. | | Deposit date: | 2017-07-05 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Deconstruction of the Ras switching cycle through saturation mutagenesis.
Elife, 6, 2017
|
|
3EER
 
 | | High resolution structure of putative organic hydroperoxide resistance protein from Vibrio cholerae O1 biovar eltor str. N16961 | | Descriptor: | ACETATE ION, IMIDAZOLE, Organic hydroperoxide resistance protein, ... | | Authors: | Nocek, B, Maltseva, N, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-09-05 | | Release date: | 2008-10-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | High resolution crystal structure of the organic hydroperoxide
resistance protein from Vibrio cholerae O1 biovar eltor str. N16961
To be Published
|
|
5WDS
 
 | | Choanoflagellate Salpingoeca rosetta Ras with GDP bound | | Descriptor: | GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kondo, Y, Gee, C.L, Kuriyan, J. | | Deposit date: | 2017-07-05 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Deconstruction of the Ras switching cycle through saturation mutagenesis.
Elife, 6, 2017
|
|
5WDP
 
 | | H-Ras mutant L120A bound to GMP-PNP at 277K | | Descriptor: | CALCIUM ION, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Cofsky, J.C, Bandaru, P, Gee, C.L, Kuriyan, J. | | Deposit date: | 2017-07-05 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Deconstruction of the Ras switching cycle through saturation mutagenesis.
Elife, 6, 2017
|
|
5WDR
 
 | | Choanoflagellate Salpingoeca rosetta Ras with GMP-PNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras protein, ... | | Authors: | Kondo, Y, Gee, C.L, Kuriyan, J. | | Deposit date: | 2017-07-05 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Deconstruction of the Ras switching cycle through saturation mutagenesis.
Elife, 6, 2017
|
|
4ZIL
 
 | | Crystal structure of Rv2466c and oxidoreductase from Mycobacterium tuberculosis in its reduced state | | Descriptor: | DSBA oxidoreductase, MAGNESIUM ION | | Authors: | Albesa-Jove, D, Urresti, S, Comino, N, Tersa, M, Guerin, M.E. | | Deposit date: | 2015-04-28 | | Release date: | 2015-11-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.507 Å) | | Cite: | The Redox State Regulates the Conformation of Rv2466c to Activate the Antitubercular Prodrug TP053.
J.Biol.Chem., 290, 2015
|
|
4ZDJ
 
 | | Crystal structure of the M. tuberculosis CTP synthase PyrG in complex with two UTP molecules | | Descriptor: | CTP synthase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Bellinzoni, M, Barilone, N, Alzari, P.M. | | Deposit date: | 2015-04-17 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Thiophenecarboxamide Derivatives Activated by EthA Kill Mycobacterium tuberculosis by Inhibiting the CTP Synthetase PyrG.
Chem.Biol., 22, 2015
|
|
4ZDI
 
 | |
4ZDK
 
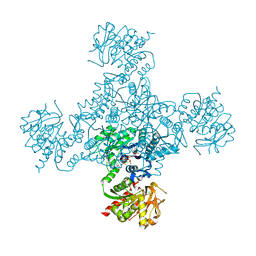 | | Crystal structure of the M. tuberculosis CTP synthase PyrG in complex with UTP, AMP-PCP and oxonorleucine | | Descriptor: | 5-OXO-L-NORLEUCINE, CTP synthase, MAGNESIUM ION, ... | | Authors: | Bellinzoni, M, Barilone, N, Alzari, P.M. | | Deposit date: | 2015-04-17 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Thiophenecarboxamide Derivatives Activated by EthA Kill Mycobacterium tuberculosis by Inhibiting the CTP Synthetase PyrG.
Chem.Biol., 22, 2015
|
|
1HVX
 
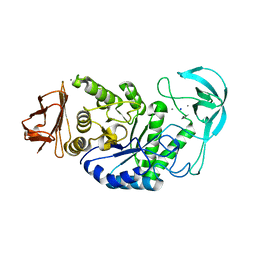 | | BACILLUS STEAROTHERMOPHILUS ALPHA-AMYLASE | | Descriptor: | ALPHA-AMYLASE, CALCIUM ION, SODIUM ION | | Authors: | Suvd, D, Fujimoto, Z, Takase, K, Matsumura, M, Mizuno, H. | | Deposit date: | 2001-01-08 | | Release date: | 2001-01-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Bacillus stearothermophilus alpha-amylase: possible factors determining the thermostability.
J.Biochem., 129, 2001
|
|
2MT7
 
 | |
2OKW
 
 | |
3USB
 
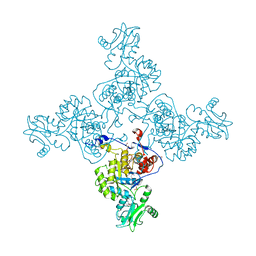 | | Crystal Structure of Bacillus anthracis Inosine Monophosphate Dehydrogenase in the complex with IMP | | Descriptor: | CHLORIDE ION, GLYCEROL, INOSINIC ACID, ... | | Authors: | Kim, Y, Zhang, R, Wu, R, Gu, M, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-11-23 | | Release date: | 2011-12-07 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Bacillus anthracis inosine 5'-monophosphate dehydrogenase in action: the first bacterial series of structures of phosphate ion-, substrate-, and product-bound complexes.
Biochemistry, 51, 2012
|
|
5K10
 
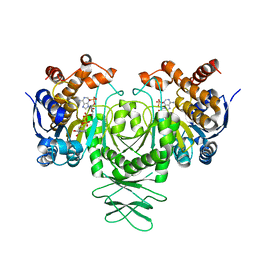 | | Cryo-EM structure of isocitrate dehydrogenase (IDH1) | | Descriptor: | Isocitrate dehydrogenase [NADP] cytoplasmic, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Merk, A, Bartesaghi, A, Banerjee, S, Falconieri, V, Rao, P, Earl, L, Milne, J, Subramaniam, S. | | Deposit date: | 2016-05-17 | | Release date: | 2016-06-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Breaking Cryo-EM Resolution Barriers to Facilitate Drug Discovery.
Cell, 165, 2016
|
|
5K11
 
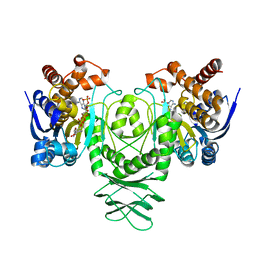 | | Cryo-EM structure of isocitrate dehydrogenase (IDH1) in inhibitor-bound state | | Descriptor: | Isocitrate dehydrogenase [NADP] cytoplasmic, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Merk, A, Bartesaghi, A, Banerjee, S, Falconieri, V, Rao, P, Earl, L, Milne, J, Subramaniam, S. | | Deposit date: | 2016-05-17 | | Release date: | 2016-06-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Breaking Cryo-EM Resolution Barriers to Facilitate Drug Discovery.
Cell, 165, 2016
|
|
5K12
 
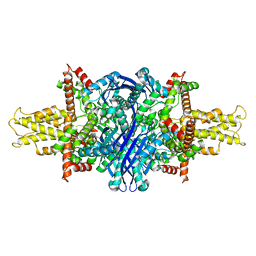 | | Cryo-EM structure of glutamate dehydrogenase at 1.8 A resolution | | Descriptor: | Glutamate dehydrogenase 1, mitochondrial | | Authors: | Merk, A, Bartesaghi, A, Banerjee, S, Falconieri, V, Rao, P, Earl, L, Milne, J, Subramaniam, S. | | Deposit date: | 2016-05-17 | | Release date: | 2016-06-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (1.8 Å) | | Cite: | Breaking Cryo-EM Resolution Barriers to Facilitate Drug Discovery.
Cell, 165, 2016
|
|
6JYG
 
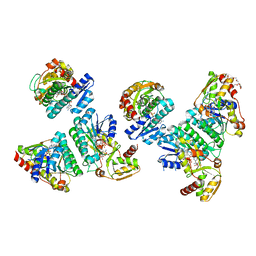 | | Crystal Structure of L-threonine dehydrogenase from Phytophthora infestans | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, CITRATE ANION, L-threonine 3-dehydrogenase, ... | | Authors: | Yoneda, K, Sakuraba, H, Ohshima, T. | | Deposit date: | 2019-04-26 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Catalytic properties and crystal structure of UDP-galactose 4-epimerase-like l-threonine 3-dehydrogenase from Phytophthora infestans.
Enzyme.Microb.Technol., 140, 2020
|
|
7UAD
 
 | | Crystal structure of human PTPN2 with inhibitor ABBV-CLS-484 | | Descriptor: | 5-{(7R)-1-fluoro-3-hydroxy-7-[(3-methylbutyl)amino]-5,6,7,8-tetrahydronaphthalen-2-yl}-1lambda~6~,2,5-thiadiazolidine-1,1,3-trione, Tyrosine-protein phosphatase non-receptor type 2 | | Authors: | Longenecker, K.L, Qiu, W, Sun, Q, Frost, J.M. | | Deposit date: | 2022-03-12 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.044 Å) | | Cite: | The PTPN2/PTPN1 inhibitor ABBV-CLS-484 unleashes potent anti-tumour immunity.
Nature, 622, 2023
|
|
6ARO
 
 | |
6AOX
 
 | |
6AS8
 
 | |
