3AQP
 
 | | Crystal structure of SecDF, a translocon-associated membrane protein, from Thermus thrmophilus | | Descriptor: | Probable SecDF protein-export membrane protein | | Authors: | Tsukazaki, T, Mori, H, Echizen, Y, Ishitani, R, Fukai, S, Tanaka, T, Perederina, A, Vassylyev, D.G, Kohno, T, Ito, K, Nureki, O. | | Deposit date: | 2010-11-16 | | Release date: | 2011-05-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure and function of a membrane component SecDF that enhances protein export
Nature, 474, 2011
|
|
3AQO
 
 | |
5CZZ
 
 | | Crystal structure of Staphylococcus aureus Cas9 in complex with sgRNA and target DNA (TTGAAT PAM) | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9, DNA (28-MER), ... | | Authors: | Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2015-08-01 | | Release date: | 2015-09-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Staphylococcus aureus Cas9.
Cell, 162, 2015
|
|
4MKP
 
 | | Crystal structure of human cGAS apo form | | Descriptor: | Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Kato, K, Ishii, R, Ishitani, R, Nureki, O. | | Deposit date: | 2013-09-05 | | Release date: | 2013-10-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.953 Å) | | Cite: | Structural and Functional Analyses of DNA-Sensing and Immune Activation by Human cGAS
Plos One, 8, 2013
|
|
4XJ3
 
 | | Crystal structure of Vibrio cholerae DncV GTP bound form | | Descriptor: | 1,2-ETHANEDIOL, Cyclic AMP-GMP synthase, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Kato, K, Ishii, R, Ishitani, R, Nureki, O. | | Deposit date: | 2015-01-08 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Basis for the Catalytic Mechanism of DncV, Bacterial Homolog of Cyclic GMP-AMP Synthase
Structure, 23, 2015
|
|
4XJ1
 
 | | Crystal structure of Vibrio cholerae DncV apo form | | Descriptor: | 1,2-ETHANEDIOL, Cyclic AMP-GMP synthase | | Authors: | Kato, K, Ishii, R, Ishitani, R, Nureki, O. | | Deposit date: | 2015-01-08 | | Release date: | 2015-04-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural Basis for the Catalytic Mechanism of DncV, Bacterial Homolog of Cyclic GMP-AMP Synthase
Structure, 23, 2015
|
|
4XJ4
 
 | | Crystal structure of Vibrio cholerae DncV 3'-deoxy ATP bound form | | Descriptor: | 1,2-ETHANEDIOL, 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, Cyclic AMP-GMP synthase, ... | | Authors: | Kato, K, Ishii, R, Ishitani, R, Nureki, O. | | Deposit date: | 2015-01-08 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.596 Å) | | Cite: | Structural Basis for the Catalytic Mechanism of DncV, Bacterial Homolog of Cyclic GMP-AMP Synthase
Structure, 23, 2015
|
|
4XJ5
 
 | | Crystal structure of Vibrio cholerae DncV 3'-deoxy GTP bound form | | Descriptor: | 1,2-ETHANEDIOL, 3'-DEOXY-GUANOSINE-5'-TRIPHOSPHATE, Cyclic AMP-GMP synthase, ... | | Authors: | Kato, K, Ishii, R, Ishitani, R, Nureki, O. | | Deposit date: | 2015-01-08 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.552 Å) | | Cite: | Structural Basis for the Catalytic Mechanism of DncV, Bacterial Homolog of Cyclic GMP-AMP Synthase
Structure, 23, 2015
|
|
8JMS
 
 | |
4YZI
 
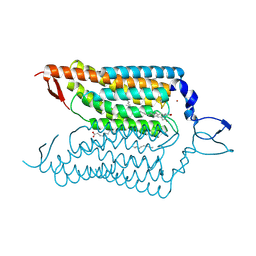 | | Crystal structure of blue-shifted channelrhodopsin mutant (T198G/G202A) | | Descriptor: | OLEIC ACID, RETINAL, Sensory opsin A,Archaeal-type opsin 2, ... | | Authors: | Kato, H.E, Kamiya, M, Ishitani, R, Hayashi, S, Nureki, O. | | Deposit date: | 2015-03-25 | | Release date: | 2015-05-27 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Atomistic design of microbial opsin-based blue-shifted optogenetics tools.
Nat Commun, 6, 2015
|
|
1EG7
 
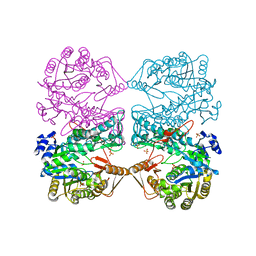 | | THE CRYSTAL STRUCTURE OF FORMYLTETRAHYDROFOLATE SYNTHETASE FROM MOORELLA THERMOACETICA | | Descriptor: | FORMYLTETRAHYDROFOLATE SYNTHETASE, SULFATE ION | | Authors: | Radfar, R, Shin, R, Sheldrick, G.M, Minor, W, Lovell, C.R, Odom, J.D, Dunlap, R.B, Lebioda, L. | | Deposit date: | 2000-02-14 | | Release date: | 2001-02-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of N(10)-formyltetrahydrofolate synthetase from Moorella thermoacetica.
Biochemistry, 39, 2000
|
|
5EGH
 
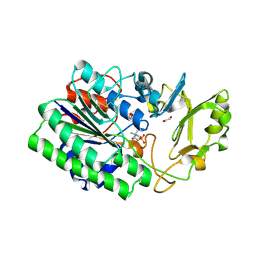 | | Structure of ENPP6, a choline-specific glycerophosphodiester-phosphodiesterase in complex with phosphocholine | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Morita, J, Kano, K, Kato, K, Takita, H, Ishitani, R, Nishimasu, H, Nureki, O, Aoki, J. | | Deposit date: | 2015-10-27 | | Release date: | 2016-03-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Structure and biological function of ENPP6, a choline-specific glycerophosphodiester-phosphodiesterase
Sci Rep, 6, 2016
|
|
5EGE
 
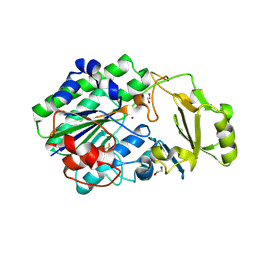 | | Structure of ENPP6, a choline-specific glycerophosphodiester-phosphodiesterase | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Morita, J, Kano, K, Kato, K, Takita, H, Ishitani, R, Nishimasu, H, Nureki, O, Aoki, J. | | Deposit date: | 2015-10-27 | | Release date: | 2016-03-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and biological function of ENPP6, a choline-specific glycerophosphodiester-phosphodiesterase
Sci Rep, 6, 2016
|
|
4P79
 
 | | Crystal structure of mouse claudin-15 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Claudin-15 | | Authors: | Suzuki, H, Nishizawa, T, Tani, K, Yamazaki, Y, Tamura, A, Ishitani, R, Dohmae, N, Tsukita, S, Nureki, O, Fujiyoshi, Y. | | Deposit date: | 2014-03-26 | | Release date: | 2014-04-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a claudin provides insight into the architecture of tight junctions.
Science, 344, 2014
|
|
3G5Q
 
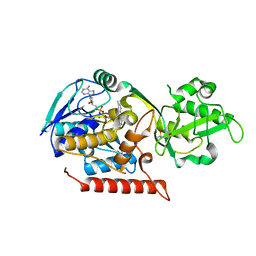 | | Crystal structure of Thermus thermophilus TrmFO | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, FLAVIN-ADENINE DINUCLEOTIDE, Methylenetetrahydrofolate--tRNA-(uracil-5-)-methyltransferase trmFO | | Authors: | Nishimasu, H, Ishitani, R, Hori, H, Nureki, O. | | Deposit date: | 2009-02-05 | | Release date: | 2009-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Atomic structure of a folate/FAD-dependent tRNA T54 methyltransferase
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3G5S
 
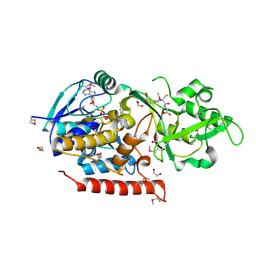 | | Crystal structure of Thermus thermophilus TrmFO in complex with glutathione | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, GLUTATHIONE, ... | | Authors: | Nishimasu, H, Ishitani, R, Hori, H, Nureki, O. | | Deposit date: | 2009-02-05 | | Release date: | 2009-05-19 | | Last modified: | 2011-12-14 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Atomic structure of a folate/FAD-dependent tRNA T54 methyltransferase
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3G5R
 
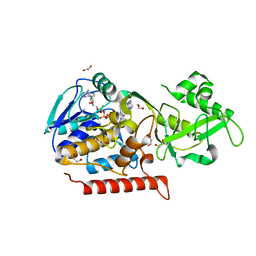 | | Crystal structure of Thermus thermophilus TrmFO in complex with tetrahydrofolate | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Nishimasu, H, Ishitani, R, Hori, H, Nureki, O. | | Deposit date: | 2009-02-05 | | Release date: | 2009-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Atomic structure of a folate/FAD-dependent tRNA T54 methyltransferase
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
8ZJ2
 
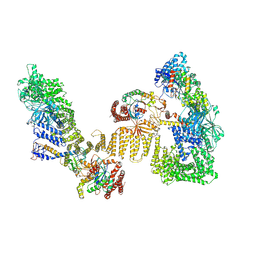 | | Cryo-EM structure of the RhoG/DOCK5/ELMO1/Rac1 complex | | Descriptor: | Dedicator of cytokinesis protein 5, Engulfment and cell motility protein 1, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Kukimoto-Niino, M, Katsura, K, Ishizuka-Katsura, Y, Mishima-Tsumagari, C, Yonemochi, M, Inoue, M, Nakagawa, R, Kaushik, R, Zhang, K.Y.J, Shirouzu, M. | | Deposit date: | 2024-05-14 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.66 Å) | | Cite: | RhoG facilitates a conformational transition in the guanine nucleotide exchange factor complex DOCK5/ELMO1 to an open state.
J.Biol.Chem., 300, 2024
|
|
8ZJL
 
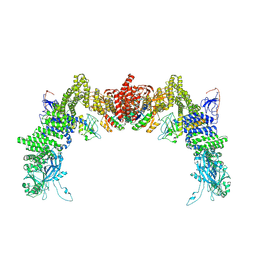 | | Structure of DOCK5/ELMO1/Rac1 core (RhoG/DOCK5/ELMO1/Rac1 dataset, class 4) | | Descriptor: | Dedicator of cytokinesis protein 5, Engulfment and cell motility protein 1, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Kukimoto-Niino, M, Katsura, K, Ishizuka-Katsura, Y, Mishima-Tsumagari, C, Yonemochi, M, Inoue, M, Nakagawa, R, Kaushik, R, Zhang, K.Y.J, Shirouzu, M. | | Deposit date: | 2024-05-15 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.31 Å) | | Cite: | RhoG facilitates a conformational transition in the guanine nucleotide exchange factor complex DOCK5/ELMO1 to an open state.
J.Biol.Chem., 300, 2024
|
|
8ZJJ
 
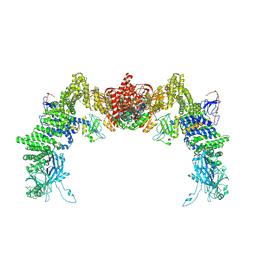 | | Structure of DOCK5/ELMO1/Rac1 core (RhoG/DOCK5/ELMO1/Rac1 dataset, class 2) | | Descriptor: | Dedicator of cytokinesis protein 5, Engulfment and cell motility protein 1, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Kukimoto-Niino, M, Katsura, K, Ishizuka-Katsura, Y, Mishima-Tsumagari, C, Yonemochi, M, Inoue, M, Nakagawa, R, Kaushik, R, Zhang, K.Y.J, Shirouzu, M. | | Deposit date: | 2024-05-15 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.23 Å) | | Cite: | RhoG facilitates a conformational transition in the guanine nucleotide exchange factor complex DOCK5/ELMO1 to an open state.
J.Biol.Chem., 300, 2024
|
|
8ZJK
 
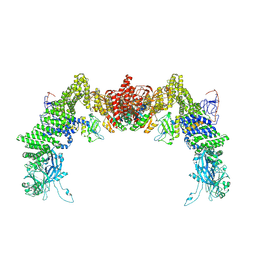 | | Structure of DOCK5/ELMO1/Rac1 core (RhoG/DOCK5/ELMO1/Rac1 dataset, class 3) | | Descriptor: | Dedicator of cytokinesis protein 5, Engulfment and cell motility protein 1, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Kukimoto-Niino, M, Katsura, K, Ishizuka-Katsura, Y, Mishima-Tsumagari, C, Yonemochi, M, Inoue, M, Nakagawa, R, Kaushik, R, Zhang, K.Y.J, Shirouzu, M. | | Deposit date: | 2024-05-15 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.23 Å) | | Cite: | RhoG facilitates a conformational transition in the guanine nucleotide exchange factor complex DOCK5/ELMO1 to an open state.
J.Biol.Chem., 300, 2024
|
|
8ZJI
 
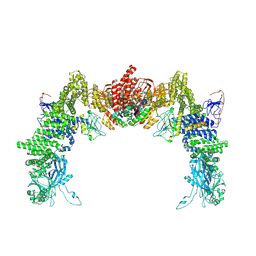 | | Structure of DOCK5/ELMO1/Rac1 core (RhoG/DOCK5/ELMO1/Rac1 dataset, class 1) | | Descriptor: | Dedicator of cytokinesis protein 5, Engulfment and cell motility protein 1, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Kukimoto-Niino, M, Katsura, K, Ishizuka-Katsura, Y, Mishima-Tsumagari, C, Yonemochi, M, Inoue, M, Nakagawa, R, Kaushik, R, Zhang, K.Y.J, Shirouzu, M. | | Deposit date: | 2024-05-15 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.23 Å) | | Cite: | RhoG facilitates a conformational transition in the guanine nucleotide exchange factor complex DOCK5/ELMO1 to an open state.
J.Biol.Chem., 300, 2024
|
|
8ZJM
 
 | | Structure of DOCK5/ELMO1/Rac1 core (RhoG/DOCK5/ELMO1/Rac1 dataset, class 5) | | Descriptor: | Dedicator of cytokinesis protein 5, Engulfment and cell motility protein 1, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Kukimoto-Niino, M, Katsura, K, Ishizuka-Katsura, Y, Mishima-Tsumagari, C, Yonemochi, M, Inoue, M, Nakagawa, R, Kaushik, R, Zhang, K.Y.J, Shirouzu, M. | | Deposit date: | 2024-05-15 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.52 Å) | | Cite: | RhoG facilitates a conformational transition in the guanine nucleotide exchange factor complex DOCK5/ELMO1 to an open state.
J.Biol.Chem., 300, 2024
|
|
8XM7
 
 | | Cryo-EM structure of the RhoG/DOCK5/ELMO1/Rac1 complex: RhoG/DOCK5/ELMO1 focused map | | Descriptor: | Dedicator of cytokinesis protein 5, Engulfment and cell motility protein 1, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Kukimoto-Niino, M, Katsura, K, Ishizuka-Katsura, Y, Mishima-Tsumagari, C, Yonemochi, M, Inoue, M, Nakagawa, R, Kaushik, R, Zhang, K.Y.J, Shirouzu, M. | | Deposit date: | 2023-12-27 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.91 Å) | | Cite: | RhoG facilitates a conformational transition in the guanine nucleotide exchange factor complex DOCK5/ELMO1 to an open state.
J.Biol.Chem., 300, 2024
|
|
3WTR
 
 | | Crystal structure of E. coli YfcM bound to Co(II) | | Descriptor: | COBALT (II) ION, Uncharacterized protein | | Authors: | Kobayashi, K, Ishii, R, Ishitani, R, Nureki, O. | | Deposit date: | 2014-04-19 | | Release date: | 2015-04-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The non-canonical hydroxylase structure of YfcM reveals a metal ion-coordination motif required for EF-P hydroxylation
Nucleic Acids Res., 42, 2014
|
|
