1JV7
 
 | | BACTERIORHODOPSIN O-LIKE INTERMEDIATE STATE OF THE D85S MUTANT AT 2.25 ANGSTROM RESOLUTION | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin, RETINAL | | Authors: | Rouhani, S, Cartailler, J.-P, Facciotti, M.T, Walian, P, Needleman, R, Lanyi, J.K, Glaeser, R.M, Luecke, H. | | Deposit date: | 2001-08-28 | | Release date: | 2001-10-31 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the D85S mutant of bacteriorhodopsin: model of an O-like photocycle intermediate.
J.Mol.Biol., 313, 2001
|
|
7Y5G
 
 | | Cryo-EM structure of a eukaryotic ZnT8 in the presence of zinc | | Descriptor: | ZINC ION, Zinc transporter 8 | | Authors: | Zhang, S, Fu, C, Luo, Y, Sun, Z, Su, Z, Zhou, X. | | Deposit date: | 2022-06-17 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Cryo-EM structure of a eukaryotic zinc transporter at a low pH suggests its Zn 2+ -releasing mechanism.
J.Struct.Biol., 215, 2022
|
|
7Y5H
 
 | | Cryo-EM structure of a eukaryotic ZnT8 at a low pH | | Descriptor: | ZINC ION, Zinc transporter 8 | | Authors: | Zhang, S, Fu, C, Luo, Y, Sun, Z, Su, Z, Zhou, X. | | Deposit date: | 2022-06-17 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Cryo-EM structure of a eukaryotic zinc transporter at a low pH suggests its Zn 2+ -releasing mechanism.
J.Struct.Biol., 215, 2022
|
|
4U1E
 
 | | Crystal structure of the eIF3b-CTD/eIF3i/eIF3g-NTD translation initiation complex | | Descriptor: | Eukaryotic translation initiation factor 3 subunit B, Eukaryotic translation initiation factor 3 subunit G, Eukaryotic translation initiation factor 3 subunit I | | Authors: | Zhang, S, Erzberger, J.P, Schaefer, T, Ban, N. | | Deposit date: | 2014-07-15 | | Release date: | 2014-09-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular Architecture of the 40SeIF1eIF3 Translation Initiation Complex.
Cell, 158, 2014
|
|
7OQ6
 
 | | Crystal structure of cytochrome P450 Sas16 from Streptomyces asterosporus | | Descriptor: | Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE, THIOCYANATE ION | | Authors: | Zhang, L, Zhang, S, Bechthold, A, Einsle, O. | | Deposit date: | 2021-06-02 | | Release date: | 2022-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | P450-mediated dehydrotyrosine formation during WS9326 biosynthesis proceeds via dehydrogenation of a specific acylated dipeptide substrate.
Acta Pharm Sin B, 13, 2023
|
|
6QMQ
 
 | | NF-YB/C Heterodimer in Complex with NF-YA-derived Peptide Stabilized with C8-Hydrocarbon Linker | | Descriptor: | Nuclear transcription factor Y subunit alpha, Nuclear transcription factor Y subunit beta, Nuclear transcription factor Y subunit gamma, ... | | Authors: | Kiehstaller, S, Jeganathan, S, Pearce, N.M, Wendt, M, Grossmann, T.N, Hennig, S. | | Deposit date: | 2019-02-08 | | Release date: | 2019-10-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Constrained Peptides with Fine-Tuned Flexibility Inhibit NF-Y Transcription Factor Assembly.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
6QMP
 
 | | NF-YB/C Heterodimer in Complex with NF-YA Peptide | | Descriptor: | Nuclear transcription factor Y subunit alpha, Nuclear transcription factor Y subunit beta, Nuclear transcription factor Y subunit gamma | | Authors: | Kiehstaller, S, Jeganathan, S, Pearce, N.M, Wendt, M, Grossmann, T.N, Hennig, S. | | Deposit date: | 2019-02-08 | | Release date: | 2019-10-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Constrained Peptides with Fine-Tuned Flexibility Inhibit NF-Y Transcription Factor Assembly.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
6QMS
 
 | | NF-YB/C Heterodimer in Complex with NF-YA-derived Peptide Stabilized with C11-Hydrocarbon Linker | | Descriptor: | Nuclear transcription factor Y subunit alpha, Nuclear transcription factor Y subunit beta, Nuclear transcription factor Y subunit gamma | | Authors: | Kiehstaller, S, Jeganathan, S, Pearce, N.M, Wendt, M, Grossmann, T.N, Hennig, S. | | Deposit date: | 2019-02-08 | | Release date: | 2019-10-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Constrained Peptides with Fine-Tuned Flexibility Inhibit NF-Y Transcription Factor Assembly.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
6QW2
 
 | | The Transcriptional Regulator PrfA-A218G mutant from Listeria Monocytogenes | | Descriptor: | ISOPROPYL ALCOHOL, Listeriolysin positive regulatory factor A, SODIUM ION | | Authors: | Hall, M, Grundstrom, C, Hansen, S, Brannstrom, K, Johansson, J, Sauer-Eriksson, A.E. | | Deposit date: | 2019-03-05 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Novel Growth-Based Selection Strategy Identifies New Constitutively Active Variants of the Major Virulence Regulator PrfA in Listeria monocytogenes.
J.Bacteriol., 202, 2020
|
|
6QWF
 
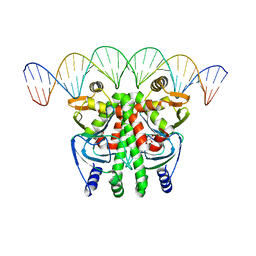 | | The Transcriptional Regulator PrfA-A94V mutant from Listeria Monocytogenes in complex with a 30-bp operator PrfA-box motif | | Descriptor: | DNA (30-MER), GLYCEROL, Listeriolysin positive regulatory factor A | | Authors: | Hall, M, Grundstrom, C, Hansen, S, Brannstrom, K, Johansson, J, Sauer-Eriksson, A.E. | | Deposit date: | 2019-03-05 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Novel Growth-Based Selection Strategy Identifies New Constitutively Active Variants of the Major Virulence Regulator PrfA in Listeria monocytogenes.
J.Bacteriol., 202, 2020
|
|
6QVY
 
 | | The Transcriptional Regulator PrfA-A94V mutant from Listeria Monocytogenes | | Descriptor: | ISOPROPYL ALCOHOL, Listeriolysin regulatory protein, SODIUM ION | | Authors: | Hall, M, Grundstrom, C, Hansen, S, Brannstrom, K, Johansson, J, Sauer-Eriksson, A.E. | | Deposit date: | 2019-03-05 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.686 Å) | | Cite: | A Novel Growth-Based Selection Strategy Identifies New Constitutively Active Variants of the Major Virulence Regulator PrfA in Listeria monocytogenes.
J.Bacteriol., 202, 2020
|
|
6QWK
 
 | | The Transcriptional Regulator PrfA-L140F mutant from Listeria Monocytogenes in complex with a 30-bp operator PrfA-box motif | | Descriptor: | DNA (30-MER), GLYCEROL, Listeriolysin positive regulatory factor A | | Authors: | Hall, M, Grundstrom, C, Hansen, S, Brannstrom, K, Johansson, J, Sauer-Eriksson, A.E. | | Deposit date: | 2019-03-05 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A Novel Growth-Based Selection Strategy Identifies New Constitutively Active Variants of the Major Virulence Regulator PrfA in Listeria monocytogenes.
J.Bacteriol., 202, 2020
|
|
6QVZ
 
 | | The Transcriptional Regulator PrfA-L140H mutant from Listeria Monocytogenes | | Descriptor: | ISOPROPYL ALCOHOL, Listeriolysin positive regulatory factor A, SODIUM ION | | Authors: | Hall, M, Grundstrom, C, Hansen, S, Brannstrom, K, Johansson, J, Sauer-Eriksson, A.E. | | Deposit date: | 2019-03-05 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.705 Å) | | Cite: | A Novel Growth-Based Selection Strategy Identifies New Constitutively Active Variants of the Major Virulence Regulator PrfA in Listeria monocytogenes.
J.Bacteriol., 202, 2020
|
|
6QWH
 
 | | The Transcriptional Regulator PrfA-L140H mutant from Listeria Monocytogenes in complex with a 30-bp operator PrfA-box motif | | Descriptor: | DNA (30-MER), GLYCEROL, Listeriolysin positive regulatory factor A | | Authors: | Hall, M, Grundstrom, C, Hansen, S, Brannstrom, K, Johansson, J, Sauer-Eriksson, A.E. | | Deposit date: | 2019-03-05 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A Novel Growth-Based Selection Strategy Identifies New Constitutively Active Variants of the Major Virulence Regulator PrfA in Listeria monocytogenes.
J.Bacteriol., 202, 2020
|
|
5UIS
 
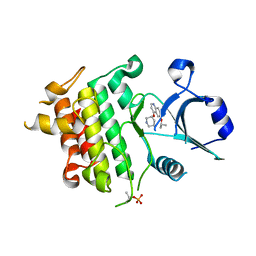 | | Crystal structure of IRAK4 in complex with compound 12 | | Descriptor: | 4-{[(3R)-piperidin-3-yl]oxy}-6-[(propan-2-yl)oxy]quinoline-7-carboxamide, Interleukin-1 receptor-associated kinase 4 | | Authors: | Han, S, Chang, J.S. | | Deposit date: | 2017-01-14 | | Release date: | 2017-05-24 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of Clinical Candidate 1-{[(2S,3S,4S)-3-Ethyl-4-fluoro-5-oxopyrrolidin-2-yl]methoxy}-7-methoxyisoquinoline-6-carboxamide (PF-06650833), a Potent, Selective Inhibitor of Interleukin-1 Receptor Associated Kinase 4 (IRAK4), by Fragment-Based Drug Design.
J. Med. Chem., 60, 2017
|
|
5UIQ
 
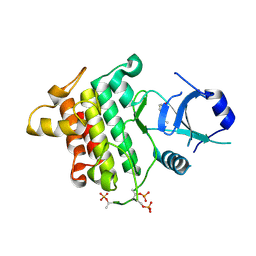 | | Crystal structure of IRAK4 in complex with compound 9 | | Descriptor: | 2-[(propan-2-yl)oxy]benzamide, Interleukin-1 receptor-associated kinase 4 | | Authors: | Han, S, Chang, J.S. | | Deposit date: | 2017-01-14 | | Release date: | 2017-05-24 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Discovery of Clinical Candidate 1-{[(2S,3S,4S)-3-Ethyl-4-fluoro-5-oxopyrrolidin-2-yl]methoxy}-7-methoxyisoquinoline-6-carboxamide (PF-06650833), a Potent, Selective Inhibitor of Interleukin-1 Receptor Associated Kinase 4 (IRAK4), by Fragment-Based Drug Design.
J. Med. Chem., 60, 2017
|
|
5UIR
 
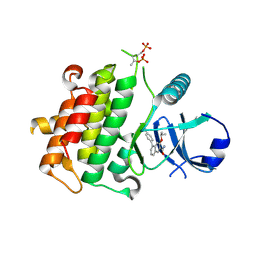 | | Crystal structure of IRAK4 in complex with compound 11 | | Descriptor: | 5-(4-cyanophenyl)-3-[(propan-2-yl)oxy]naphthalene-2-carboxamide, Interleukin-1 receptor-associated kinase 4 | | Authors: | Han, S, Chang, J.S. | | Deposit date: | 2017-01-14 | | Release date: | 2017-05-24 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Discovery of Clinical Candidate 1-{[(2S,3S,4S)-3-Ethyl-4-fluoro-5-oxopyrrolidin-2-yl]methoxy}-7-methoxyisoquinoline-6-carboxamide (PF-06650833), a Potent, Selective Inhibitor of Interleukin-1 Receptor Associated Kinase 4 (IRAK4), by Fragment-Based Drug Design.
J. Med. Chem., 60, 2017
|
|
5UIU
 
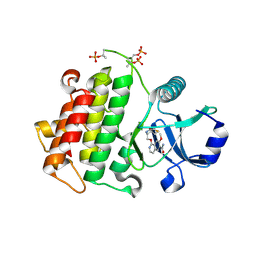 | | Crystal structure of IRAK4 in complex with compound 30 | | Descriptor: | 1-{[(2S,3S,4S)-3-ethyl-4-fluoro-5-oxopyrrolidin-2-yl]methoxy}-7-methoxyisoquinoline-6-carboxamide, Interleukin-1 receptor-associated kinase 4 | | Authors: | Han, S, Chang, J.S. | | Deposit date: | 2017-01-14 | | Release date: | 2017-05-24 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Discovery of Clinical Candidate 1-{[(2S,3S,4S)-3-Ethyl-4-fluoro-5-oxopyrrolidin-2-yl]methoxy}-7-methoxyisoquinoline-6-carboxamide (PF-06650833), a Potent, Selective Inhibitor of Interleukin-1 Receptor Associated Kinase 4 (IRAK4), by Fragment-Based Drug Design.
J. Med. Chem., 60, 2017
|
|
5UIT
 
 | | Crystal structure of IRAK4 in complex with compound 14 | | Descriptor: | 1-{[(2S)-5-oxopyrrolidin-2-yl]methoxy}-7-[(propan-2-yl)oxy]isoquinoline-6-carboxamide, Interleukin-1 receptor-associated kinase 4 | | Authors: | Han, S, Chang, J.S. | | Deposit date: | 2017-01-14 | | Release date: | 2017-05-24 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Discovery of Clinical Candidate 1-{[(2S,3S,4S)-3-Ethyl-4-fluoro-5-oxopyrrolidin-2-yl]methoxy}-7-methoxyisoquinoline-6-carboxamide (PF-06650833), a Potent, Selective Inhibitor of Interleukin-1 Receptor Associated Kinase 4 (IRAK4), by Fragment-Based Drug Design.
J. Med. Chem., 60, 2017
|
|
6MNY
 
 | |
7CN8
 
 | | Cryo-EM structure of PCoV_GX spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein, ... | | Authors: | Wang, X, Yu, J, Zhang, S, Qiao, S, Zeng, J, Tian, L. | | Deposit date: | 2020-07-30 | | Release date: | 2021-03-03 | | Last modified: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Bat and pangolin coronavirus spike glycoprotein structures provide insights into SARS-CoV-2 evolution.
Nat Commun, 12, 2021
|
|
7CN4
 
 | | Cryo-EM structure of bat RaTG13 spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, X, Zhang, S, Qiao, S, Yu, J, Zeng, J, Tian, L. | | Deposit date: | 2020-07-30 | | Release date: | 2021-03-03 | | Last modified: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Bat and pangolin coronavirus spike glycoprotein structures provide insights into SARS-CoV-2 evolution.
Nat Commun, 12, 2021
|
|
3T9T
 
 | | Crystal structure of BTK mutant (F435T,K596R) complexed with Imidazo[1,5-a]quinoxaline | | Descriptor: | (2Z)-4-(dimethylamino)-N-{7-fluoro-4-[(2-methylphenyl)amino]imidazo[1,5-a]quinoxalin-8-yl}-N-methylbut-2-enamide, GLYCEROL, Tyrosine-protein kinase ITK/TSK | | Authors: | Han, S, Caspers, N. | | Deposit date: | 2011-08-03 | | Release date: | 2011-10-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Imidazo[1,5-a]quinoxalines as irreversible BTK inhibitors for the treatment of rheumatoid arthritis.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
4U1F
 
 | |
5C26
 
 | | Crystal structure of SYK in complex with compound 1 | | Descriptor: | 3-{8-[(3,4-dimethoxyphenyl)amino]imidazo[1,2-a]pyrazin-6-yl}benzamide, GLU-VAL-PTR-GLU-SER-PRO, Tyrosine-protein kinase SYK | | Authors: | Han, S, Chang, J. | | Deposit date: | 2015-06-15 | | Release date: | 2015-10-07 | | Last modified: | 2016-02-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Imidazotriazines: Spleen Tyrosine Kinase (Syk) Inhibitors Identified by Free-Energy Perturbation (FEP).
Chemmedchem, 11, 2016
|
|
