8BGC
 
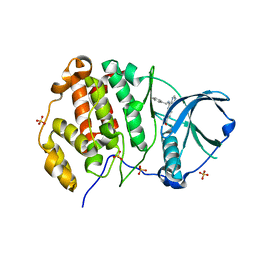 | | Crystal structure of human Casein Kinase II subunit alpha (CK2a1) in complex with compound 2 (AA-CS-9-003) | | Descriptor: | 5-[(phenylmethyl)amino]pyrimido[4,5-c]quinoline-8-carboxylic acid, Casein kinase II subunit alpha, SULFATE ION | | Authors: | Kraemer, A, Axtman, A.D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-10-27 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of a Potent and Selective Naphthyridine-Based Chemical Probe for Casein Kinase 2.
Acs Med.Chem.Lett., 14, 2023
|
|
8B2T
 
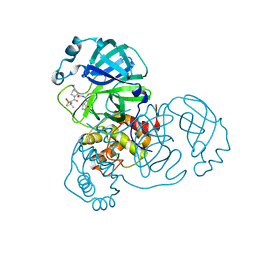 | | SARS-CoV-2 Main Protease (Mpro) in complex with nirmatrelvir alkyne | | Descriptor: | 3C-like proteinase nsp5, Nirmatrelvir (reacted form) | | Authors: | Owen, C.D, Crawshaw, A.D, Warren, A.J, Trincao, J, Zhao, Y, Brewitz, L, Malla, T.R, Salah, E, Petra, L, Strain-Damerell, C, Schofield, C.J, Walsh, M.A. | | Deposit date: | 2022-09-14 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.893 Å) | | Cite: | Alkyne Derivatives of SARS-CoV-2 Main Protease Inhibitors Including Nirmatrelvir Inhibit by Reacting Covalently with the Nucleophilic Cysteine.
J.Med.Chem., 66, 2023
|
|
2JMR
 
 | | NMR structure of the E. coli type 1 pilus subunit FimF | | Descriptor: | fimF | | Authors: | Gossert, A.D, Bettendorff, P, Puorger, C, Vetsch, M, Herrmann, T, Fiorito, F, Hiller, S, Glockshuber, R, Wuthrich, K. | | Deposit date: | 2006-11-29 | | Release date: | 2007-10-30 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the Escherichia coli type 1 pilus subunit FimF and its interactions with other pilus subunits.
J.Mol.Biol., 375, 2008
|
|
2JQM
 
 | | Yellow Fever Envelope Protein Domain III NMR Structure (S288-K398) | | Descriptor: | Envelope protein E | | Authors: | Volk, D.E, Gandham, S.H, May, F.J, Anderson, A, Barrett, A.D, Gorenstein, D.G. | | Deposit date: | 2007-06-03 | | Release date: | 2008-06-10 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Structure of yellow fever virus envelope protein domain III.
Virology, 394, 2009
|
|
8A1E
 
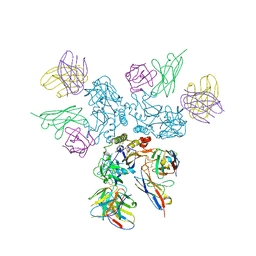 | | Rabies virus glycoprotein in complex with Fab fragments of 17C7 and 1112-1 neutralizing antibodies | | Descriptor: | Fab 1112-1 heavy chain variable domain, Fab 1112-1 light chain variable domain, Fab 17C7 heavy chain variable domain, ... | | Authors: | Ng, W.M, Fedosyuk, S, English, S, Augusto, G, Berg, A, Thorley, L, Haselon, A.S, Segireddy, R.R, Bowden, T.A, Douglas, A.D. | | Deposit date: | 2022-06-01 | | Release date: | 2022-08-17 | | Last modified: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structure of trimeric pre-fusion rabies virus glycoprotein in complex with two protective antibodies.
Cell Host Microbe, 30, 2022
|
|
2JLN
 
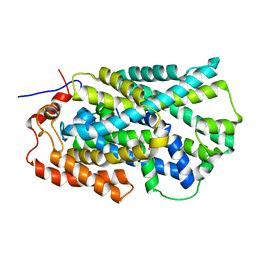 | | Structure of Mhp1, a nucleobase-cation-symport-1 family transporter | | Descriptor: | MERCURY (II) ION, MHP1, SODIUM ION | | Authors: | Weyand, S, Shimamura, T, Yajima, S, Suzuki, S, Mirza, O, Krusong, K, Carpenter, E.P, Rutherford, N.G, Hadden, J.M, O'Reilly, J, Ma, P, Saidijam, M, Patching, S.G, Hope, R.J, Norbertczak, H.T, Roach, P.C.J, Iwata, S, Henderson, P.J.F, Cameron, A.D. | | Deposit date: | 2008-09-11 | | Release date: | 2008-10-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure and Molecular Mechanism of a Nucleobase-Cation-Symport-1 Family Transporter.
Science, 322, 2008
|
|
2LS5
 
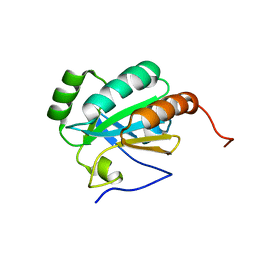 | | Solution structure of a putative protein disulfide isomerase from Bacteroides thetaiotaomicron | | Descriptor: | Uncharacterized protein | | Authors: | Harris, R, Bandaranayake, A.D, Banu, R, Bonanno, J.B, Calarese, D.A, Celikgil, A, Chamala, S, Chan, M.K, Chaparro, R, Evans, B, Garforth, S, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Lim, S, Love, J, Matikainen, B, Patel, H, Seidel, R.D, Smith, B, Stead, M, Girvin, M.E, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-04-20 | | Release date: | 2012-05-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a putative protein disulfide isomerase from Bacteroides thetaiotaomicron
To be Published
|
|
2JSB
 
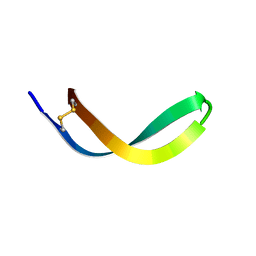 | | Solution structure of arenicin-1 | | Descriptor: | Arenicin-1 | | Authors: | Jakovkin, I.B, Hecht, O, Gelhaus, C, Krasnosdembskaya, A.D, Fedders, H, Leippe, M, Groetzinger, J. | | Deposit date: | 2007-07-02 | | Release date: | 2008-02-05 | | Last modified: | 2020-02-19 | | Method: | SOLUTION NMR | | Cite: | Structure and mode of action of the antimicrobial peptide arenicin
Biochem.J., 410, 2008
|
|
2M96
 
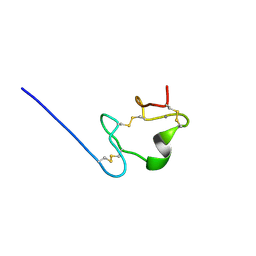 | |
2OPY
 
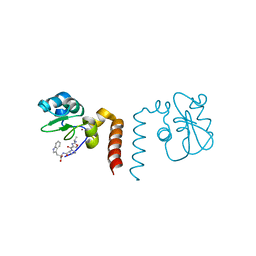 | | Smac mimic bound to BIR3-XIAP | | Descriptor: | 1-({2-[(1S)-1-AMINOETHYL]-1,3-OXAZOL-4-YL}CARBONYL)-L-PROLYL-L-TRYPTOPHAN, Baculoviral IAP repeat-containing protein 4, ZINC ION | | Authors: | Wist, A.D, Lu, G, Riedl, S.J, Shi, Y, McLendon, G.L. | | Deposit date: | 2007-01-30 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-activity based study of the Smac-binding pocket within the BIR3 domain of XIAP.
Bioorg.Med.Chem., 15, 2007
|
|
2OPZ
 
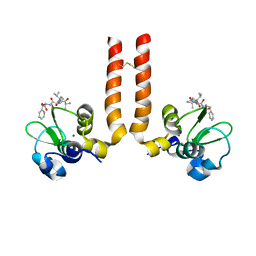 | | AVPF bound to BIR3-XIAP | | Descriptor: | AVPF (Smac homologue, N-terminal tetrapeptide), Baculoviral IAP repeat-containing protein 4, ... | | Authors: | Wist, A.D. | | Deposit date: | 2007-01-30 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-activity based study of the Smac-binding pocket within the BIR3 domain of XIAP.
Bioorg.Med.Chem., 15, 2007
|
|
2PE6
 
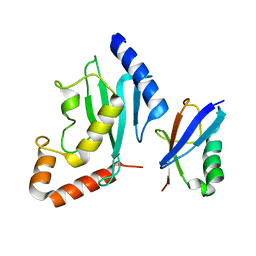 | | Non-covalent complex between human SUMO-1 and human Ubc9 | | Descriptor: | SUMO-conjugating enzyme UBC9, Small ubiquitin-related modifier 1 | | Authors: | Capili, A.D, Lima, C.D. | | Deposit date: | 2007-04-02 | | Release date: | 2007-04-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and Analysis of a Complex between SUMO and Ubc9 Illustrates Features of a Conserved E2-Ubl Interaction.
J.Mol.Biol., 369, 2007
|
|
2QTR
 
 | |
2QX9
 
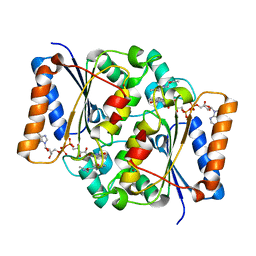 | | Crystal Structure of Quinone Reductase II | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, N-[2-(2-iodo-5-methoxy-1H-indol-3-yl)ethyl]acetamide, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Calamini, B, Santarsiero, B.D, Boutin, J.A, Mesecar, A.D. | | Deposit date: | 2007-08-10 | | Release date: | 2008-09-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Kinetic, thermodynamic and X-ray structural insights into the interaction of melatonin and analogues with quinone reductase 2.
Biochem.J., 413, 2008
|
|
2QTM
 
 | |
2P4V
 
 | | Crystal structure of the transcript cleavage factor, GreB at 2.6A resolution | | Descriptor: | Transcription elongation factor greB | | Authors: | Vassylyeva, M.N, Svetlov, V, Dearborn, A.D, Klyuyev, S, Artsimovitch, I, Vassylyev, D.G. | | Deposit date: | 2007-03-13 | | Release date: | 2008-01-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The carboxy-terminal coiled-coil of the RNA polymerase beta'-subunit is the main binding site for Gre factors.
Embo Rep., 8, 2007
|
|
6MDR
 
 | | Cryo-EM structure of the Ceru+32/GFP-17 protomer | | Descriptor: | Ceru+32, GFP-17 | | Authors: | Simon, A.J, Zhou, Y, Ramasubramani, V, Glaser, J, Pothukuchy, A, Golihar, J, Gerberich, J.C, Leggere, J.C, Morrow, B.R, Jung, C, Glotzer, S.C, Taylor, D.W, Ellington, A.D. | | Deposit date: | 2018-09-05 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Supercharging enables organized assembly of synthetic biomolecules.
Nat Chem, 11, 2019
|
|
2Q7R
 
 | | Crystal structure of human FLAP with an iodinated analog of MK-591 | | Descriptor: | 3-[3-(3,3-DIMETHYLBUTANOYL)-1-(4-IODOBENZYL)-5-(QUINOLIN-2-YLMETHOXY)-1H-INDOL-2-YL]-2,2-DIMETHYLPROPANOIC ACID, Arachidonate 5-lipoxygenase-activating protein | | Authors: | Ferguson, A.D. | | Deposit date: | 2007-06-07 | | Release date: | 2007-08-21 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Crystal structure of inhibitor-bound human 5-lipoxygenase-activating protein.
Science, 317, 2007
|
|
2QIO
 
 | |
2QTN
 
 | | Crystal Structure of Nicotinate Mononucleotide Adenylyltransferase | | Descriptor: | GLYCEROL, MAGNESIUM ION, NICOTINATE MONONUCLEOTIDE, ... | | Authors: | Sershon, V.C, Santarsiero, B.D, Mesecar, A.D. | | Deposit date: | 2007-08-02 | | Release date: | 2008-07-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Kinetic and X-ray structural evidence for negative cooperativity in substrate binding to nicotinate mononucleotide adenylyltransferase (NMAT) from Bacillus anthracis.
J.Mol.Biol., 385, 2009
|
|
6MSF
 
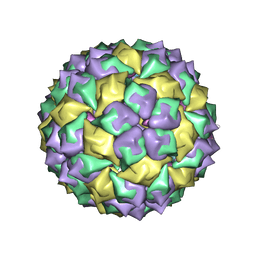 | | F6 APTAMER MS2 COAT PROTEIN COMPLEX | | Descriptor: | PROTEIN (MS2 PROTEIN CAPSID), RNA (5'-R(*CP*AP*GP*UP*CP*AP*CP*UP*GP*G)-3'), RNA (5'-R(*CP*CP*AP*CP*AP*GP*UP*CP*AP*CP*UP*GP*GP*G)-3') | | Authors: | Convery, M.A, Rowsell, S, Stonehouse, N.J, Ellington, A.D, Hirao, I, Murray, J.B, Peabody, D.S, Phillips, S.E.V, Stockley, P.G. | | Deposit date: | 1998-01-06 | | Release date: | 1998-07-08 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of an RNA aptamer-protein complex at 2.8 A resolution.
Nat.Struct.Biol., 5, 1998
|
|
6MUY
 
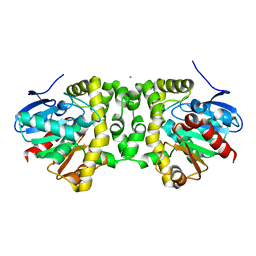 | | Fluoroacetate dehalogenase, room temperature structure solved by serial 3 degree oscillation crystallography | | Descriptor: | CALCIUM ION, Fluoroacetate dehalogenase | | Authors: | Finke, A.D, Wierman, J.L, Pare-Labrosse, O, Sarrachini, A, Besaw, J, Mehrabi, P, Gruner, S.M, Miller, R.J.D. | | Deposit date: | 2018-10-24 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fixed-target serial oscillation crystallography at room temperature.
IUCrJ, 6, 2019
|
|
2QX8
 
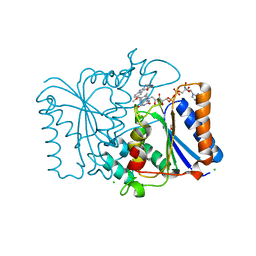 | | Crystal Structure of Quinone Reductase II | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Calamini, B, Santarsiero, B.D, Boutin, J.A, Mesecar, A.D. | | Deposit date: | 2007-08-10 | | Release date: | 2008-09-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Kinetic, thermodynamic and X-ray structural insights into the interaction of melatonin and analogues with quinone reductase 2.
Biochem.J., 413, 2008
|
|
2QX6
 
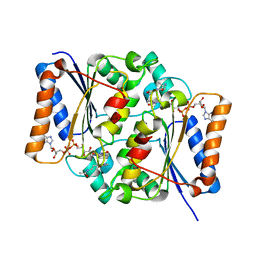 | | Crystal Structure of Quinone Reductase II | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, N-[2-(5-methoxy-1H-indol-3-yl)ethyl]acetamide, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Calamini, B, Santarsiero, B.D, Boutin, J.A, Mesecar, A.D. | | Deposit date: | 2007-08-10 | | Release date: | 2008-09-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Kinetic, thermodynamic and X-ray structural insights into the interaction of melatonin and analogues with quinone reductase 2.
Biochem.J., 413, 2008
|
|
2QWX
 
 | | Crystal Structure of Quinone Reductase II | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, N-[2-(5-methoxy-1H-indol-3-yl)ethyl]acetamide, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Calamini, B, Santarsiero, B.D, Boutin, J.A, Mesecar, A.D. | | Deposit date: | 2007-08-10 | | Release date: | 2008-04-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Kinetic, thermodynamic and X-ray structural insights into the interaction of melatonin and analogues with quinone reductase 2.
Biochem.J., 413, 2008
|
|
