4K8Y
 
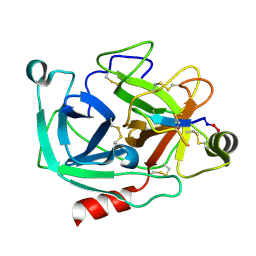 | | Atomic resolution crystal structures of Kallikrein-Related Peptidase 4 complexed with Sunflower Trypsin Inhibitor (SFTI-1) | | Descriptor: | Kallikrein-4, Trypsin inhibitor 1 | | Authors: | Ilyichova, O.V, Swedberg, J.E, de Veer, S.J, Sit, K.C, Harris, J.M, Buckle, A.M. | | Deposit date: | 2013-04-19 | | Release date: | 2014-04-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Direct and indirect mechanisms of KLK4 inhibition revealed by structure and dynamics
Sci Rep, 6, 2016
|
|
1CKQ
 
 | |
1CL8
 
 | |
4KM5
 
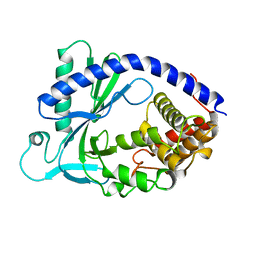 | | X-ray crystal structure of human cyclic GMP-AMP synthase (cGAS) | | Descriptor: | Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Kranzusch, P.J, Lee, A.S.Y, Berger, J.M, Doudna, J.A. | | Deposit date: | 2013-05-08 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Structure of Human cGAS Reveals a Conserved Family of Second-Messenger Enzymes in Innate Immunity.
Cell Rep, 3, 2013
|
|
4KEL
 
 | | Atomic resolution crystal structure of Kallikrein-Related Peptidase 4 complexed with a modified SFTI inhibitor FCQR(N) | | Descriptor: | Kallikrein-4, Trypsin inhibitor 1 | | Authors: | Ilyichova, O.V, Swedberg, J.E, de Veer, S.J, Sit, K.C, Harris, J.M, Buckle, A.M. | | Deposit date: | 2013-04-25 | | Release date: | 2014-04-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.148 Å) | | Cite: | KLK4 Inhibition by Cyclic and Acyclic Peptides: Structural and Dynamical Insights into Standard-Mechanism Protease Inhibitors.
Biochemistry, 58, 2019
|
|
4M1Y
 
 | | Crystal Structure of small molecule vinylsulfonamide 15 covalently bound to K-Ras G12C | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, K-Ras GTPase, N-{1-[N-(5,7-dichloro-2,1,3-benzothiadiazol-4-yl)glycyl]piperidin-4-yl}ethanesulfonamide | | Authors: | Ostrem, J.M, Peters, U, Sos, M.L, Wells, J.A, Shokat, K.M. | | Deposit date: | 2013-08-04 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.491 Å) | | Cite: | K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions.
Nature, 503, 2013
|
|
3W39
 
 | | Crystal structure of HLA-B*5201 in complexed with HIV immunodominant epitope (TAFTIPSI) | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-52 alpha chain, ... | | Authors: | Yagita, Y, Kuse, N, Kuroki, K, Gatanaga, H, Carlson, J.M, Chikata, T, Brumme, Z.L, Murakoshi, H, Akahoshi, T, Pfeifer, N, Mallal, S, John, M, Ose, T, Matsubara, H, Kanda, R, Fukunaga, Y, Honda, K, Kawashima, Y, Ariumi, Y, Oka, S, Maenaka, K, Takiguchi, M. | | Deposit date: | 2012-12-13 | | Release date: | 2013-02-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Distinct HIV-1 Escape Patterns Selected by Cytotoxic T Cells with Identical Epitope Specificity
J.Virol., 87, 2013
|
|
1D3B
 
 | | CRYSTAL STRUCTURE OF THE D3B SUBCOMPLEX OF THE HUMAN CORE SNRNP DOMAIN AT 2.0A RESOLUTION | | Descriptor: | CITRIC ACID, GLYCEROL, PROTEIN (SMALL NUCLEAR RIBONUCLEOPROTEIN ASSOCIATED PROTEIN B), ... | | Authors: | Kambach, C, Walke, S, Avis, J.M, De La Fortelle, E, Li, J, Nagai, K. | | Deposit date: | 1998-12-22 | | Release date: | 1999-12-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs.
Cell(Cambridge,Mass.), 96, 1999
|
|
1D3Y
 
 | | STRUCTURE OF THE DNA TOPOISOMERASE VI A SUBUNIT | | Descriptor: | DNA TOPOISOMERASE VI A SUBUNIT, MAGNESIUM ION, SODIUM ION | | Authors: | Nichols, M.D, DeAngelis, K.A, Keck, J.L, Berger, J.M. | | Deposit date: | 1999-10-01 | | Release date: | 1999-11-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and function of an archaeal topoisomerase VI subunit with homology to the meiotic recombination factor Spo11.
EMBO J., 18, 1999
|
|
4LPL
 
 | | Structure of CBM32-1 from a family 31 glycoside hydrolase from Clostridium perfringens | | Descriptor: | CALCIUM ION, Glycosyl hydrolase, family 31/fibronectin type III domain protein, ... | | Authors: | Grondin, J.M, Duan, D, Heather, F.S, Spencer, C.A, Allingham, J.S, Smith, S.P. | | Deposit date: | 2013-07-16 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Diverse modes of galacto-specific carbohydrate recognition by a family 31 glycoside hydrolase from Clostridium perfringens.
Plos One, 12, 2017
|
|
1D5F
 
 | | STRUCTURE OF E6AP: INSIGHTS INTO UBIQUITINATION PATHWAY | | Descriptor: | E6AP HECT CATALYTIC DOMAIN, E3 LIGASE | | Authors: | Huang, L, Kinnucan, E, Wang, G, Beaudenon, S, Howley, P.M, Huibregtse, J.M, Pavletich, N.P. | | Deposit date: | 1999-10-07 | | Release date: | 1999-11-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of an E6AP-UbcH7 complex: insights into ubiquitination by the E2-E3 enzyme cascade.
Science, 286, 1999
|
|
4JO8
 
 | | Crystal structure of the activating Ly49H receptor in complex with m157 (G1F strain) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Killer cell lectin-like receptor 8, M157 | | Authors: | Berry, R, Ng, N, Saunders, P.M, Vivian, J.P, Lin, J, Deuss, F.A, Corbett, A.J, Forbes, C.A, Widjaja, J.M, Sullivan, L.C, McAlister, A.D, Perugini, M.A, Call, M.J, Scalzo, A.A, Degli-Esposti, M.A, Coudert, J.D, Beddoe, T, Brooks, A.G, Rossjohn, J. | | Deposit date: | 2013-03-18 | | Release date: | 2013-05-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Targeting of a natural killer cell receptor family by a viral immunoevasin
Nat.Immunol., 14, 2013
|
|
1D6F
 
 | | CHALCONE SYNTHASE C164A MUTANT | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHALCONE SYNTHASE, SULFATE ION | | Authors: | Jez, J.M, Ferrer, J.L, Bowman, M.E, Dixon, R.A, Noel, J.P. | | Deposit date: | 1999-10-13 | | Release date: | 2000-02-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Dissection of malonyl-coenzyme A decarboxylation from polyketide formation in the reaction mechanism of a plant polyketide synthase.
Biochemistry, 39, 2000
|
|
4JXW
 
 | |
3UJB
 
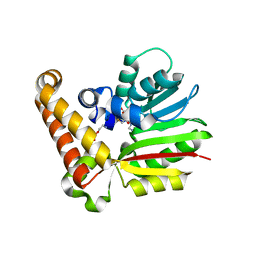 | | Phosphoethanolamine methyltransferase from Plasmodium falciparum in complex with SAH and phosphoethanolamine | | Descriptor: | PHOSPHORIC ACID MONO-(2-AMINO-ETHYL) ESTER, Phosphoethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Lee, S.G, Kim, Y, Alpert, T.D, Nagata, A, Jez, J.M. | | Deposit date: | 2011-11-07 | | Release date: | 2011-11-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.521 Å) | | Cite: | Structure and reaction mechanism of phosphoethanolamine methyltransferase from the malaria parasite Plasmodium falciparum: an antiparasitic drug target.
J.Biol.Chem., 287, 2012
|
|
3USQ
 
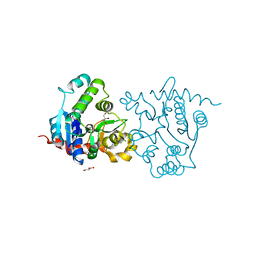 | | Structure of D159S/Y194F glycogenin mutant truncated at residue 270 | | Descriptor: | CHLORIDE ION, GLYCEROL, Glycogenin-1 | | Authors: | Issoglio, F.M, Carrizo, M.E, Romero, J.M, Curtino, J.A. | | Deposit date: | 2011-11-23 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanisms of monomeric and dimeric glycogenin autoglucosylation.
J.Biol.Chem., 287, 2012
|
|
4J9Z
 
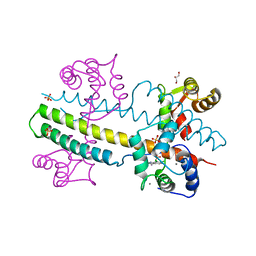 | | Calcium-calmodulin complexed with the calmodulin binding domain from a small conductance potassium channel splice variant and NS309 | | Descriptor: | (3E)-6,7-dichloro-3-(hydroxyimino)-1,3-dihydro-2H-indol-2-one, CALCIUM ION, Calmodulin, ... | | Authors: | Zhang, M, Pascal, J.M, Zhang, J.-F. | | Deposit date: | 2013-02-17 | | Release date: | 2013-03-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Unstructured to structured transition of an intrinsically disordered protein peptide in coupling Ca2+-sensing and SK channel activation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3UC1
 
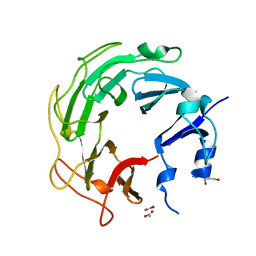 | | Mycobacterium tuberculosis gyrase type IIA topoisomerase C-terminal domain | | Descriptor: | ACETATE ION, CALCIUM ION, DNA gyrase subunit A, ... | | Authors: | Tretter, E.M, Berger, J.M. | | Deposit date: | 2011-10-25 | | Release date: | 2012-03-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Mechanisms for Defining Supercoiling Set Point of DNA Gyrase Orthologs: II. THE SHAPE OF THE GyrA SUBUNIT C-TERMINAL DOMAIN (CTD) IS NOT A SOLE DETERMINANT FOR CONTROLLING SUPERCOILING EFFICIENCY.
J.Biol.Chem., 287, 2012
|
|
3UEH
 
 | | Crystal structure of human Survivin H80A mutant | | Descriptor: | 1,2-ETHANEDIOL, Baculoviral IAP repeat-containing protein 5, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Niedzialkowska, E, Porebski, P.J, Wang, F, Higgins, J.M, Stukenberg, P.T, Minor, W. | | Deposit date: | 2011-10-30 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres.
Mol Biol Cell, 23, 2012
|
|
3UJ6
 
 | | SeMet Phosphoethanolamine methyltransferase from Plasmodium falciparum in complex with SAM and PO4 | | Descriptor: | PHOSPHATE ION, Phosphoethanolamine N-methyltransferase, S-ADENOSYLMETHIONINE | | Authors: | Lee, S.G, Kim, Y, Alpert, T.D, Nagata, A, Jez, J.M. | | Deposit date: | 2011-11-07 | | Release date: | 2011-11-30 | | Last modified: | 2012-03-21 | | Method: | X-RAY DIFFRACTION (1.974 Å) | | Cite: | Structure and reaction mechanism of phosphoethanolamine methyltransferase from the malaria parasite Plasmodium falciparum: an antiparasitic drug target.
J.Biol.Chem., 287, 2012
|
|
3UJD
 
 | | Phosphoethanolamine methyltransferase mutant (Y19F) from Plasmodium falciparum in complex with phosphocholine | | Descriptor: | PHOSPHOCHOLINE, Phosphoethanolamine N-methyltransferase | | Authors: | Lee, S.G, Kim, Y, Alpert, T.D, Nagata, A, Jez, J.M. | | Deposit date: | 2011-11-07 | | Release date: | 2011-11-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and reaction mechanism of phosphoethanolamine methyltransferase from the malaria parasite Plasmodium falciparum: an antiparasitic drug target.
J.Biol.Chem., 287, 2012
|
|
3UK3
 
 | | Crystal structure of ZNF217 bound to DNA | | Descriptor: | 5'-D(*AP*AP*TP*GP*CP*AP*GP*AP*AP*TP*CP*GP*AP*TP*TP*CP*TP*GP*CP*A)-3', 5'-D(*TP*TP*TP*GP*CP*AP*GP*AP*AP*TP*CP*GP*AP*TP*TP*CP*TP*GP*CP*A)-3', CHLORIDE ION, ... | | Authors: | Vandevenne, M.S, Jacques, D.A, Guss, J.M, Mackay, J.P. | | Deposit date: | 2011-11-08 | | Release date: | 2012-12-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Rediscovering DNA recognition by classical Zinc Fingers.
To be Published
|
|
4LPK
 
 | | Crystal Structure of K-Ras WT, GDP-bound | | Descriptor: | CALCIUM ION, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Ostrem, J.M, Peters, U, Sos, M.L, Wells, J.A, Shokat, K.M. | | Deposit date: | 2013-07-16 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions.
Nature, 503, 2013
|
|
4LXM
 
 | | Crystal Structure of Human Beta Secretase in Complex with compound 12a | | Descriptor: | (1S,3S,4S,5R)-3-{4-amino-3-fluoro-5-[(1,1,1,3,3,3-hexafluoropropan-2-yl)oxy]benzyl}-5-[(3-tert-butylbenzyl)amino]tetrahydro-2H-thiopyran-4-ol 1-oxide, Beta-secretase 1 | | Authors: | Rondeau, J.M, Bourgier, E. | | Deposit date: | 2013-07-30 | | Release date: | 2013-08-28 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of cyclic sulfoxide hydroxyethylamines as potent and selective beta-site APP-cleaving enzyme 1 (BACE1) inhibitors: structure based design and in vivo reduction of amyloid beta-peptides.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4LYH
 
 | | Crystal Structure of small molecule vinylsulfonamide 9 covalently bound to K-Ras G12C | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, N-{1-[N-(4-chloro-5-iodo-2-methoxyphenyl)glycyl]piperidin-4-yl}ethanesulfonamide, ... | | Authors: | Ostrem, J.M, Peters, U, Sos, M.L, Wells, J.A, Shokat, K.M. | | Deposit date: | 2013-07-31 | | Release date: | 2013-11-27 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.371 Å) | | Cite: | K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions.
Nature, 503, 2013
|
|
