7VSF
 
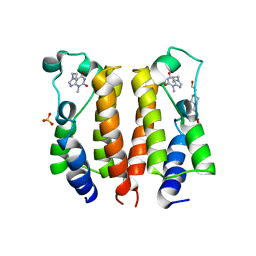 | | crystal structure of BRD2-BD1 in complex with purine derivative | | Descriptor: | 3-methyl-7-propyl-purine-2,6-dione, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Bromodomain-containing protein 2, ... | | Authors: | Padmanabhan, B, Arole, A, Deshmukh, P, Ashok, S, Mathur, S. | | Deposit date: | 2021-10-26 | | Release date: | 2023-02-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and biochemical insights into purine-based drug molecules in hBRD2 delineate a unique binding mode opening new vistas in the design of inhibitors of the BET family.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7VS0
 
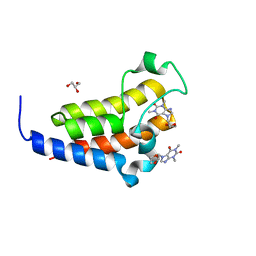 | | crystal structure of BRD2-BD2 in complex with purine derivative | | Descriptor: | Bromodomain-containing protein 2, Doxofylline, GLYCEROL | | Authors: | Padmanabhan, B, Arole, A, Deshmukh, P, Ashok, S, Mathur, S. | | Deposit date: | 2021-10-25 | | Release date: | 2023-02-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural and biochemical insights into purine-based drug molecules in hBRD2 delineate a unique binding mode opening new vistas in the design of inhibitors of the BET family.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7VRK
 
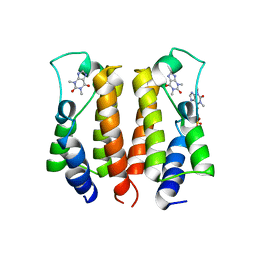 | | crystal structure of BRD2-BD1 in complex with purine derivative | | Descriptor: | Bromodomain-containing protein 2, SULFATE ION, THEOPHYLLINE | | Authors: | Padmanabhan, B, Arole, A, Deshmukh, P, Ashok, S, Mathur, S. | | Deposit date: | 2021-10-23 | | Release date: | 2023-02-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural and biochemical insights into purine-based drug molecules in hBRD2 delineate a unique binding mode opening new vistas in the design of inhibitors of the BET family.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7VRH
 
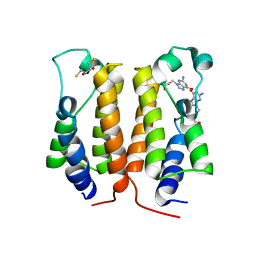 | | crystal structure of BRD2-BD1 in complex with guanosine analog | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 9-HYROXYETHOXYMETHYLGUANINE, Bromodomain-containing protein 2, ... | | Authors: | Padmanabhan, B, Arole, A, Deshmukh, P, Ashok, S, Mathur, S. | | Deposit date: | 2021-10-22 | | Release date: | 2023-02-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and biochemical insights into purine-based drug molecules in hBRD2 delineate a unique binding mode opening new vistas in the design of inhibitors of the BET family.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7VRI
 
 | | crystal structure of BRD2-BD2 in complex with guanosine analog | | Descriptor: | 9-HYROXYETHOXYMETHYLGUANINE, Bromodomain-containing protein 2 | | Authors: | Padmanabhan, B, Arole, A, Deshmukh, P, Ashok, S, Mathur, S. | | Deposit date: | 2021-10-23 | | Release date: | 2023-02-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and biochemical insights into purine-based drug molecules in hBRD2 delineate a unique binding mode opening new vistas in the design of inhibitors of the BET family.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7VS1
 
 | | crystal structure of BRD2-BD2 in complex with purine derivative | | Descriptor: | 3-methyl-7-propyl-purine-2,6-dione, Bromodomain-containing protein 2, GLYCEROL | | Authors: | Padmanabhan, B, Arole, A, Deshmukh, P, Ashok, S, Mathur, S. | | Deposit date: | 2021-10-25 | | Release date: | 2023-02-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural and biochemical insights into purine-based drug molecules in hBRD2 delineate a unique binding mode opening new vistas in the design of inhibitors of the BET family.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7VRZ
 
 | | crystal structure of BRD2-BD1 in complex with purine derivative | | Descriptor: | Bromodomain-containing protein 2, Doxofylline, SULFATE ION | | Authors: | Padmanabhan, B, Arole, A, Deshmukh, P, Ashok, S, Mathur, S. | | Deposit date: | 2021-10-25 | | Release date: | 2023-02-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural and biochemical insights into purine-based drug molecules in hBRD2 delineate a unique binding mode opening new vistas in the design of inhibitors of the BET family.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
5OLU
 
 | | The crystal structure of a highly thermostable carboxyl esterase from Bacillus coagulans in complex with glycerol | | Descriptor: | ACETATE ION, Alpha/beta hydrolase family protein, CHLORIDE ION, ... | | Authors: | Gourlay, L.J, Nakhnoukh, C, Bolognesi, M. | | Deposit date: | 2017-07-28 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A stereospecific carboxyl esterase from Bacillus coagulans hosting nonlipase activity within a lipase-like fold.
FEBS J., 285, 2018
|
|
1WBF
 
 | | WINGED BEAN LECTIN, SACCHARIDE FREE FORM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, MANGANESE (II) ION, ... | | Authors: | Manoj, N, Srinivas, V.R, Suguna, K. | | Deposit date: | 1998-12-16 | | Release date: | 1999-12-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of basic winged-bean lectin and a comparison with its saccharide-bound form.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
5M6A
 
 | | Crystal structure of cardiotoxic Bence-Jones light chain dimer H9 | | Descriptor: | Bence-Jones light chain, GLYCEROL, PHOSPHATE ION | | Authors: | Oberti, L, Rognoni, P, Bacarizo, J, Bolognesi, M, Ricagno, S. | | Deposit date: | 2016-10-24 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Concurrent structural and biophysical traits link with immunoglobulin light chains amyloid propensity.
Sci Rep, 7, 2017
|
|
5M76
 
 | | Crystal structure of cardiotoxic Bence-Jones light chain dimer H10 | | Descriptor: | BROMIDE ION, light chain dimer | | Authors: | Oberti, L, Rognoni, P, Bacarizo, J, Bolognesi, M, Ricagno, S. | | Deposit date: | 2016-10-26 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Concurrent structural and biophysical traits link with immunoglobulin light chains amyloid propensity.
Sci Rep, 7, 2017
|
|
4L3C
 
 | | Structure of HLA-A2 in complex with D76N b2m mutant and NY-ESO1 double mutant | | Descriptor: | Beta-2-microglobulin, CHLORIDE ION, GLYCEROL, ... | | Authors: | Halabelian, L, Giorgetti, S, Bellotti, V, Bolognesi, M, Ricagno, S. | | Deposit date: | 2013-06-05 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Class I Major Histocompatibility Complex, the Trojan Horse for Secretion of Amyloidogenic beta 2-Microglobulin.
J.Biol.Chem., 289, 2014
|
|
5M6I
 
 | | Crystal structure of non-cardiotoxic Bence-Jones light chain dimer M8 | | Descriptor: | SODIUM ION, light chain dimer | | Authors: | Oberti, L, Rognoni, P, Russo, R, Bacarizo, J, Bolognesi, M, Ricagno, S. | | Deposit date: | 2016-10-25 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Concurrent structural and biophysical traits link with immunoglobulin light chains amyloid propensity.
Sci Rep, 7, 2017
|
|
4L29
 
 | | Structure of wtMHC class I with NY-ESO1 double mutant | | Descriptor: | Beta-2-microglobulin, CHLORIDE ION, GLYCEROL, ... | | Authors: | Halabelian, L, Giorgetti, S, Bellotti, V, Bolognesi, M, Ricagno, S. | | Deposit date: | 2013-06-04 | | Release date: | 2013-12-25 | | Last modified: | 2014-02-26 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Class I Major Histocompatibility Complex, the Trojan Horse for Secretion of Amyloidogenic beta 2-Microglobulin.
J.Biol.Chem., 289, 2014
|
|
5MUH
 
 | | Crystal structure of an amyloidogenic light chain dimer H7 | | Descriptor: | light chain dimer | | Authors: | Oberti, L, Rognoni, P, Russo, R, Maritan, M, Bacarizo, J, Bolognesi, M, Ricagno, S. | | Deposit date: | 2017-01-13 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Concurrent structural and biophysical traits link with immunoglobulin light chains amyloid propensity.
Sci Rep, 7, 2017
|
|
5MVG
 
 | | Crystal structure of non-amyloidogenic light chain dimer M7 | | Descriptor: | GLYCEROL, light chain dimer | | Authors: | Oberti, L, Rognoni, P, Bacarizo, J, Bolognesi, M, Ricagno, S. | | Deposit date: | 2017-01-16 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Concurrent structural and biophysical traits link with immunoglobulin light chains amyloid propensity.
Sci Rep, 7, 2017
|
|
5MTL
 
 | | Crystal structure of an amyloidogenic light chain | | Descriptor: | light chain dimer,IGL@ protein,IGL@ protein | | Authors: | Oberti, L, Rognoni, P, Russo, R, Bacarizo, J, Bolognesi, M, Ricagno, S. | | Deposit date: | 2017-01-10 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Concurrent structural and biophysical traits link with immunoglobulin light chains amyloid propensity.
Sci Rep, 7, 2017
|
|
5MUD
 
 | | Crystal structure of an amyloidogenic light chain dimer H6 | | Descriptor: | light chain dimer,IGL@ protein | | Authors: | Oberti, L, Bacarizo, J, Maritan, M, Rognoni, P, Bolognesi, M, Ricagno, S. | | Deposit date: | 2017-01-13 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Concurrent structural and biophysical traits link with immunoglobulin light chains amyloid propensity.
Sci Rep, 7, 2017
|
|
7ZS6
 
 | | Crystal structure of Apis mellifera RidA | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Reactive intermediate deaminase A, ... | | Authors: | Visentin, C, Rizzi, G, Ricagno, S. | | Deposit date: | 2022-05-06 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Apis mellifera RidA, a novel member of the canonical YigF/YER057c/UK114 imine deiminase superfamily of enzymes pre-empting metabolic damage.
Biochem.Biophys.Res.Commun., 616, 2022
|
|
2HKK
 
 | | Carbonic anhydrase activators: Solution and X-ray crystallography for the interaction of andrenaline with various carbonic anhydrase isoforms | | Descriptor: | Carbonic anhydrase 2, L-EPINEPHRINE, MERCURY (II) ION, ... | | Authors: | Temperini, C, Innocenti, A, Vullo, D, Scozzafava, A, Supuran, C.T. | | Deposit date: | 2006-07-05 | | Release date: | 2007-05-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Carbonic anhydrase activators: L-Adrenaline plugs the active site entrance of isozyme II, activating better isoforms I, IV, VA, VII, and XIV.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
4YUR
 
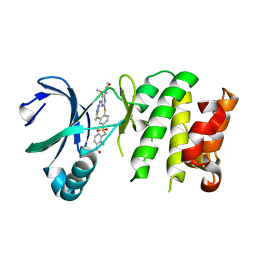 | | Crystal Structure of Plk4 Kinase Domain Bound to Centrinone | | Descriptor: | 2-({2-fluoro-4-[(2-fluoro-3-nitrobenzyl)sulfonyl]phenyl}sulfanyl)-5-methoxy-N-(3-methyl-1H-pyrazol-5-yl)-6-(morpholin-4-yl)pyrimidin-4-amine, Serine/threonine-protein kinase PLK4 | | Authors: | Shiau, A.K, Motamedi, A. | | Deposit date: | 2015-03-19 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Cell biology. Reversible centriole depletion with an inhibitor of Polo-like kinase 4.
Science, 348, 2015
|
|
6UEL
 
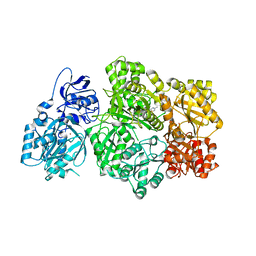 | | CPS1 bound to allosteric inhibitor H3B-193 | | Descriptor: | Carbamoyl-phosphate synthase [ammonia], mitochondrial, N~1~-[(4-fluorophenyl)methyl]-N~1~-methyl-N~4~-(4-methyl-1,3-thiazol-2-yl)piperidine-1,4-dicarboxamide, ... | | Authors: | Larsen, N.A, Nguyen, T.V. | | Deposit date: | 2019-09-21 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Small Molecule Inhibition of CPS1 Activity through an Allosteric Pocket.
Cell Chem Biol, 27, 2020
|
|
3B18
 
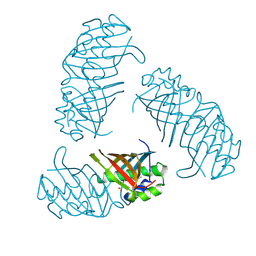 | |
6Q9Z
 
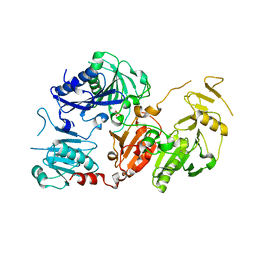 | | Crystal structure of the pathological G167R variant of calcium-free human gelsolin, | | Descriptor: | GLYCEROL, Gelsolin, SULFATE ION | | Authors: | Boni, F, Scalone, E, Milani, M, Eloise, M, de Rosa, M. | | Deposit date: | 2018-12-18 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | The structure of N184K amyloidogenic variant of gelsolin highlights the role of the H-bond network for protein stability and aggregation properties.
Eur.Biophys.J., 49, 2020
|
|
6QBF
 
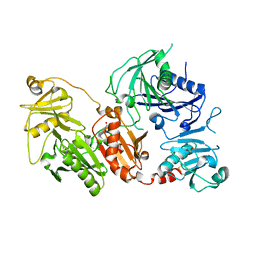 | | Crystal structure of the pathological D187N variant of calcium-free human gelsolin. | | Descriptor: | GLYCEROL, Gelsolin, SODIUM ION, ... | | Authors: | Scalone, E, Boni, F, Milani, M, Eloise, M, de Rosa, M. | | Deposit date: | 2018-12-21 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.499 Å) | | Cite: | The structure of N184K amyloidogenic variant of gelsolin highlights the role of the H-bond network for protein stability and aggregation properties.
Eur.Biophys.J., 49, 2020
|
|
