8T50
 
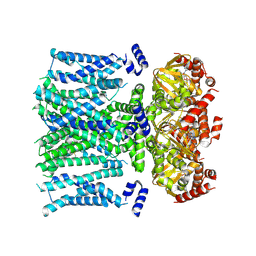 | | Open human HCN1 F186C S264C bound to cAMP, reconstituted in LMNG + SPL | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 | | Authors: | Burtscher, V, Mount, J, Cowgill, J, Chang, Y, Bickel, K, Yuan, P, Chanda, B. | | Deposit date: | 2023-06-12 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for hyperpolarization-dependent opening of human HCN1 channel.
Nat Commun, 15, 2024
|
|
8T4M
 
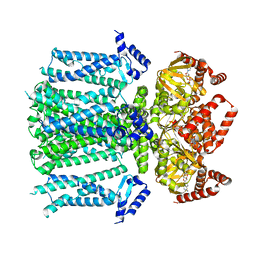 | | Closed human HCN1 F186C S264C bound to cAMP, reconstituted in LMNG + SPL | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 | | Authors: | Burtscher, V, Mount, J, Cowgill, J, Chang, Y, Bickel, K, Yuan, P, Chanda, B. | | Deposit date: | 2023-06-09 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structural basis for hyperpolarization-dependent opening of human HCN1 channel.
Nat Commun, 15, 2024
|
|
2DVW
 
 | |
2DVS
 
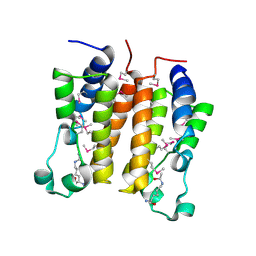 | | Crystal structure analysis of the N-terminal bromodomain of human BRD2 complexed with acetylated histone H4 peptide | | Descriptor: | bromodomain-containing protein 2, histone H4 | | Authors: | Nakamura, Y, Umehara, T, Shirouzu, M, Padmanabhan, B, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-08-01 | | Release date: | 2007-08-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural Basis for Acetylated Histone H4 Recognition by the Human BRD2 Bromodomain.
J.Biol.Chem., 285, 2010
|
|
2DVR
 
 | | Crystal structure analysis of the N-terminal bromodomain of human BRD2 complexed with acetylated histone H4 peptide | | Descriptor: | bromodomain-containing protein 2, histone H4 | | Authors: | Nakamura, Y, Umehara, T, Shirouzu, M, Padmanabhan, B, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-08-01 | | Release date: | 2007-08-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Acetylated Histone H4 Recognition by the Human BRD2 Bromodomain.
J.Biol.Chem., 285, 2010
|
|
2DVQ
 
 | | Crystal structure analysis of the N-terminal bromodomain of human BRD2 complexed with acetylated histone H4 peptide | | Descriptor: | Bromodomain-containing protein 2, histone H4 | | Authors: | Nakamura, Y, Umehara, T, Shirouzu, M, Padmanabhan, B, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-08-01 | | Release date: | 2007-08-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural Basis for Acetylated Histone H4 Recognition by the Human BRD2 Bromodomain.
J.Biol.Chem., 285, 2010
|
|
8C50
 
 | |
6DO6
 
 | | NMR solution structure of wild type apo hFABP1 at 308 K | | Descriptor: | Fatty acid-binding protein, liver | | Authors: | Scanlon, M.J, Mohanty, B, Doak, B.C, Patil, R. | | Deposit date: | 2018-06-09 | | Release date: | 2018-12-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A ligand-induced structural change in fatty acid-binding protein 1 is associated with potentiation of peroxisome proliferator-activated receptor alpha agonists.
J. Biol. Chem., 294, 2019
|
|
6DRG
 
 | | NMR solution structure of wild type hFABP1 with GW7647 | | Descriptor: | 2-[(4-{2-[(4-cyclohexylbutyl)(cyclohexylcarbamoyl)amino]ethyl}phenyl)sulfanyl]-2-methylpropanoic acid, Fatty acid-binding protein, liver | | Authors: | Scanlon, M.J, Mohanty, B, Doak, B.C, Patil, R. | | Deposit date: | 2018-06-11 | | Release date: | 2018-12-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A ligand-induced structural change in fatty acid-binding protein 1 is associated with potentiation of peroxisome proliferator-activated receptor alpha agonists.
J. Biol. Chem., 294, 2019
|
|
8GTY
 
 | |
5DG4
 
 | | Crystal structure of monomer human cellular retinol binding protein II-Y60L | | Descriptor: | ACETATE ION, Retinol-binding protein 2 | | Authors: | Assar, Z, Nossoni, Z, Wang, W, Gieger, J.H, Borhan, B. | | Deposit date: | 2015-08-27 | | Release date: | 2016-09-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Domain-Swapped Dimers of Intracellular Lipid-Binding Proteins: Evidence for Ordered Folding Intermediates.
Structure, 24, 2016
|
|
8D6H
 
 | | Q108K:K40L:T51C:T53A:R58L:Q38F:Q4F mutant of hCRBPII bound to synthetic fluorophore CM1V after UV irradiation | | Descriptor: | (2E)-3-[7-(diethylamino)-2-oxo-2H-1-benzopyran-3-yl]prop-2-enal, bound form, ACETATE ION, ... | | Authors: | Bingham, C.R, Geiger, J.H, Borhan, B. | | Deposit date: | 2022-06-06 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Light controlled reversible Michael addition of cysteine: a new tool for dynamic site-specific labeling of proteins.
Analyst, 148, 2023
|
|
8D6N
 
 | | Q108K:K40L:T53A:R58L:Q38F:Q4F mutant of hCRBPII bound to synthetic fluorophore CM1V | | Descriptor: | (2E)-3-[7-(diethylamino)-2-oxo-2H-1-benzopyran-3-yl]prop-2-enal, bound form, ACETATE ION, ... | | Authors: | Bingham, C.R, Geiger, J.H, Borhan, B. | | Deposit date: | 2022-06-06 | | Release date: | 2023-02-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Light controlled reversible Michael addition of cysteine: a new tool for dynamic site-specific labeling of proteins.
Analyst, 148, 2023
|
|
8DB2
 
 | | Q108K:K40L:T51C:T53A:R58L:Q38F mutant of hCRBPII bound to synthetic fluorophore CM1V | | Descriptor: | (2E)-3-[7-(diethylamino)-2-oxo-2H-1-benzopyran-3-yl]prop-2-enal, bound form, Retinol-binding protein 2 | | Authors: | Bingham, C.R, Geiger, J.H, Borhan, B, Staples, R. | | Deposit date: | 2022-06-14 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Light controlled reversible Michael addition of cysteine: a new tool for dynamic site-specific labeling of proteins.
Analyst, 148, 2023
|
|
8D6L
 
 | | Q108K:K40L:T51C:T53A:R58L:Q38F:Q4F mutant of hCRBPII bound to synthetic fluorophore CM1V | | Descriptor: | (2E)-3-[7-(diethylamino)-2-oxo-2H-1-benzopyran-3-yl]prop-2-enal, bound form, GLYCEROL, ... | | Authors: | Bingham, C.R, Geiger, J.H, Borhan, B. | | Deposit date: | 2022-06-06 | | Release date: | 2023-02-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Light controlled reversible Michael addition of cysteine: a new tool for dynamic site-specific labeling of proteins.
Analyst, 148, 2023
|
|
2DZO
 
 | |
2DZN
 
 | |
6DO7
 
 | | NMR solution structure of wild type hFABP1 with GW7647 | | Descriptor: | Fatty acid-binding protein, liver | | Authors: | Scanlon, M.J, Mohanty, B, Doak, B.C, Patil, R. | | Deposit date: | 2018-06-09 | | Release date: | 2019-01-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A ligand-induced structural change in fatty acid-binding protein 1 is associated with potentiation of peroxisome proliferator-activated receptor alpha agonists.
J. Biol. Chem., 294, 2019
|
|
8TJR
 
 | | CRYO-EM STRUCTURE OF HIV-1 BG505DS-SOSIP.664 ENV TRIMER BOUND TO HERH-c.01 FAB | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody HERH-a.01 Heavy Chain, ... | | Authors: | Morano, N.C, Hoyt, F, Hansen, B, Fischer, E, Shapiro, L. | | Deposit date: | 2023-07-24 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | CRYO-EM STRUCTURE OF HIV-1 BG505DS-SOSIP.664 ENV TRIMER BOUND TO HERH-c.01 FAB
To Be Published
|
|
5YGE
 
 | | ArgA complexed with AceCoA and glutamate | | Descriptor: | ACETYL COENZYME *A, Amino-acid acetyltransferase, CACODYLIC ACID, ... | | Authors: | Yang, X, Wu, L, Ran, Y, Xu, A, Zhang, B, Yang, X, Zhang, R, Rao, Z, Li, J. | | Deposit date: | 2017-09-22 | | Release date: | 2017-10-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.039 Å) | | Cite: | Crystal structure of l-glutamate N-acetyltransferase ArgA from Mycobacterium tuberculosis
Biochim. Biophys. Acta, 1865, 2017
|
|
7YZK
 
 | | Structure of Mycobacterium tuberculosis adenylyl cyclase Rv1625c / Cya | | Descriptor: | 3'-O-(N-METHYLANTHRANILOYL)-GUANOSINE-5'-TRIPHOSPHATE, Adenylate cyclase, MANGANESE (II) ION, ... | | Authors: | Mehta, V, Khanppnavar, B, Korkhov, V.M. | | Deposit date: | 2022-02-20 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Structure of Mycobacterium tuberculosis Cya, an evolutionary ancestor of the mammalian membrane adenylyl cyclases.
Elife, 11, 2022
|
|
5Z9Y
 
 | | Crystal structure of Mycobacterium tuberculosis thiazole synthase (ThiG) complexed with DXP | | Descriptor: | 1-DEOXY-D-XYLULOSE-5-PHOSPHATE, Thiazole synthase | | Authors: | Zhang, J, Zhang, B, Zhao, Y, Yang, X, Huang, M, Cui, P, Zhang, W, Li, J, Zhang, Y. | | Deposit date: | 2018-02-05 | | Release date: | 2018-04-11 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Snapshots of catalysis: Structure of covalently bound substrate trapped in Mycobacterium tuberculosis thiazole synthase (ThiG).
Biochem. Biophys. Res. Commun., 497, 2018
|
|
5NP0
 
 | | Closed dimer of human ATM (Ataxia telangiectasia mutated) | | Descriptor: | Serine-protein kinase ATM | | Authors: | Baretic, D, Pollard, H.K, Fisher, D.I, Johnson, C.M, Santhanam, B, Truman, C.M, Kouba, T, Fersht, A.R, Phillips, C, Williams, R.L. | | Deposit date: | 2017-04-13 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Structures of closed and open conformations of dimeric human ATM.
Sci Adv, 3, 2017
|
|
5NP1
 
 | | Open protomer of human ATM (Ataxia telangiectasia mutated) | | Descriptor: | Serine-protein kinase ATM | | Authors: | Baretic, D, Pollard, H.K, Fisher, D.I, Johnson, C.M, Santhanam, B, Truman, C.M, Kouba, T, Fersht, A.R, Phillips, C, Williams, R.L. | | Deposit date: | 2017-04-13 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Structures of closed and open conformations of dimeric human ATM.
Sci Adv, 3, 2017
|
|
8WDA
 
 | | Cryo-EM structure of the substrate-bound DppABCD complex | | Descriptor: | (2S)-1-(hexadecanoyloxy)propan-2-yl (10S)-10-methyloctadecanoate, PALMITIC ACID, Probable dipeptide-transport ATP-binding protein ABC transporter DppD, ... | | Authors: | Hu, T, Zhang, B, Rao, Z. | | Deposit date: | 2023-09-14 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Cryo-EM structure of the substrate-bound DppABCD complex
To Be Published
|
|
