1GFL
 
 | |
1GWF
 
 | | Compound II structure of Micrococcus Lysodeikticus catalase | | Descriptor: | ACETATE ION, Catalase, OXYGEN ATOM, ... | | Authors: | Murshudov, G.N, Grebenko, A.I, Brannigan, J.A, Antson, A.A, Barynin, V.V, Dodson, G.G, Dauter, Z, Wilson, K.S, Melik-Adamyan, W.R. | | Deposit date: | 2002-03-15 | | Release date: | 2002-04-26 | | Last modified: | 2018-06-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The Structures of Micrococcus Lysodeikticus Catalase, its Ferryl Intermediate (Compound II) and Nadph Complex.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1GWE
 
 | | Atomic resolution structure of Micrococcus Lysodeikticus catalase | | Descriptor: | Catalase, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Murshudov, G.N, Grebenko, A.I, Brannigan, J.A, Antson, A.A, Barynin, V.V, Dodson, G.G, Dauter, Z, Wilson, K.S, Melik-Adamyan, W.R. | | Deposit date: | 2002-03-15 | | Release date: | 2002-04-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (0.88 Å) | | Cite: | The Structures of Micrococcus Lysodeikticus Catalase, its Ferryl Intermediate (Compound II) and Nadph Complex.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1H4Z
 
 | | Structure of the Anti-Sigma Factor Antagonist SpoIIAA in its Unphosphorylated Form | | Descriptor: | ANTI-SIGMA F FACTOR ANTAGONIST | | Authors: | Seavers, P.R, Lewis, R.J, Brannigan, J.A, Verschueren, K.H.G, Murshudov, G.N, Wilkinson, A.J. | | Deposit date: | 2001-05-16 | | Release date: | 2001-07-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structure of the Bacillus Cell Fate Determinant Spoiiaa in Phosphorylated and Unphosphorylated Forms
Structure, 9, 2001
|
|
1H4X
 
 | | Structure of the Bacillus Cell Fate Determinant SpoIIAA in the Phosphorylated Form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ANTI-SIGMA F FACTOR ANTAGONIST | | Authors: | Seavers, P.R, Lewis, R.J, Brannigan, J.A, Verschueren, K.H.G, Murshudov, G.N, Wilkinson, A.J. | | Deposit date: | 2001-05-15 | | Release date: | 2001-07-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Structure of the Bacillus Cell Fate Determinant Spoiiaa in Phosphorylated and Unphosphorylated Forms
Structure, 9, 2001
|
|
1GWH
 
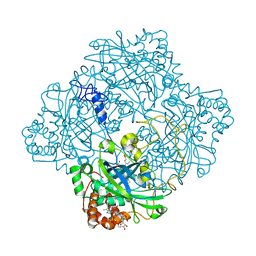 | | Atomic resolution structure of Micrococcus Lysodeikticus catalase complexed with NADPH | | Descriptor: | ACETATE ION, Catalase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Murshudov, G.N, Grebenko, A.I, Brannigan, J.A, Antson, A.A, Barynin, V.V, Dodson, G.G, Dauter, Z, Wilson, K.S, Melik-Adamyan, W.R. | | Deposit date: | 2002-03-15 | | Release date: | 2002-03-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The Structures of Micrococcus Lysodeikticus Catalase, its Ferryl Intermediate (Compound II) and Nadph Complex.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1H4Y
 
 | | Structure of the Anti-Sigma Factor Antagonist SpoIIAA in its Unphosphorylated Form | | Descriptor: | ANTI-SIGMA F FACTOR ANTAGONIST | | Authors: | Seavers, P.R, Lewis, R.J, Brannigan, J.A, Verschueren, K.H.G, Murshudov, G.N, Wilkinson, A.J. | | Deposit date: | 2001-05-16 | | Release date: | 2001-07-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structure of the Bacillus Cell Fate Determinant Spoiiaa in Phosphorylated and Unphosphorylated Forms
Structure, 9, 2001
|
|
1H9O
 
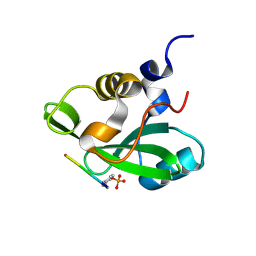 | | PHOSPHATIDYLINOSITOL 3-KINASE, P85-ALPHA SUBUNIT: C-TERMINAL SH2 DOMAIN COMPLEXED WITH A TYR751 PHOSPHOPEPTIDE FROM THE PDGF RECEPTOR, CRYSTAL STRUCTURE AT 1.79 A | | Descriptor: | BETA-PLATELET-DERIVED GROWTH FACTOR RECEPTOR, PHOSPHATIDYLINOSITOL 3-KINASE | | Authors: | Pauptit, R.A, Rowsell, S, Breeze, A.L, Murshudov, G.N, Dennis, C.A, Derbyshire, D.J, Weston, S.A. | | Deposit date: | 2001-03-14 | | Release date: | 2001-03-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | NMR Trial Models: Experiences with the Colicin Immunity Protein Im7 and the P85Alpha C-Terminal Sh2-Peptide Complex
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1HBZ
 
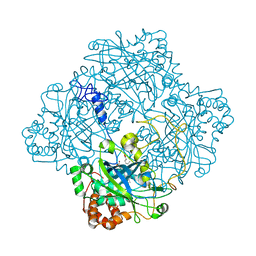 | | Catalase from Micrococcus lysodeikticu | | Descriptor: | CATALASE, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Murshudov, G.N, Grebenko, A.I, Barynin, V.V, Braningen, J, Wilson, K.S, Dauter, Z, Melik-Adamyan, W.R. | | Deposit date: | 2001-04-24 | | Release date: | 2001-05-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Three-Dimensional Structure of Catalase from Micrococcus Lysodeikticus at 1.5A Resolution
FEBS Lett., 312, 1992
|
|
4C0O
 
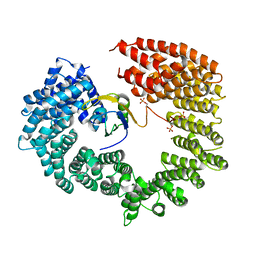 | |
4C0P
 
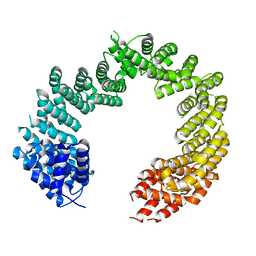 | | Unliganded Transportin 3 | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, TRANSPORTIN-3 | | Authors: | Maertens, G.N, Cook, N.J, Hare, S, Cherepanov, P. | | Deposit date: | 2013-08-06 | | Release date: | 2014-01-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural Basis for Nuclear Import of Splicing Factors by Human Transportin 3.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4DPN
 
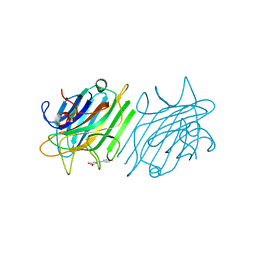 | | Crystal Structure of ConM Complexed with Resveratrol | | Descriptor: | CALCIUM ION, Concanavalin-A, MANGANESE (II) ION, ... | | Authors: | Freire, V.N, Gottfried, C, Rocha, B.A.M, Cavada, B.S, Delatorre, P, Santi-Gadelha, T, Gadelha, C.A.A, Batista, G.N, N brega, R.B, Silva-Filho, J.C. | | Deposit date: | 2012-02-13 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal Structure of ConM Complexed with Resveratrol
To be Published
|
|
4E68
 
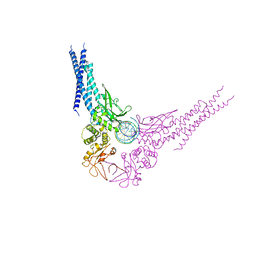 | | Unphosphorylated STAT3B core protein binding to dsDNA | | Descriptor: | DNA (5'-D(*TP*GP*CP*AP*TP*TP*TP*CP*CP*CP*GP*TP*AP*AP*AP*TP*CP*T)-3'), Signal transducer and activator of transcription 3 | | Authors: | Collie, G.W, Parkinson, G.N, Shah, R. | | Deposit date: | 2012-03-15 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.585 Å) | | Cite: | Observation of unphosphorylated STAT3 core protein binding to target dsDNA by PEMSA and X-ray crystallography.
Febs Lett., 587, 2013
|
|
4E7L
 
 | |
4E7K
 
 | | PFV integrase Target Capture Complex (TCC-Mn), freeze-trapped prior to strand transfer, at 3.0 A resolution | | Descriptor: | DNA (5'-D(*AP*TP*TP*GP*TP*CP*AP*TP*GP*GP*AP*AP*TP*TP*TP*CP*GP*CP*A)-3'), DNA (5'-D(*CP*CP*CP*GP*AP*GP*GP*CP*AP*CP*GP*TP*GP*CP*TP*AP*GP*CP*AP*CP*GP*TP*GP*CP*CP*TP*CP*GP*GP*G)-3'), DNA (5'-D(*TP*GP*CP*GP*AP*AP*AP*TP*TP*CP*CP*AP*TP*GP*AP*CP*A)-3'), ... | | Authors: | Maertens, G.N, Cherepanov, P. | | Deposit date: | 2012-03-17 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | 3'-Processing and strand transfer catalysed by retroviral integrase in crystallo.
Embo J., 31, 2012
|
|
4LZF
 
 | | A novel domain in the microcephaly protein CPAP suggests a role in centriole architecture | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Centrosomal P4.1-associated protein, SCL-interrupting locus protein homolog, ... | | Authors: | Hatzopoulos, G.N, Erat, M.C, Cutts, E, Rogala, K, Slatter, L, Stansfeld, P.J, Vakonakis, I. | | Deposit date: | 2013-07-31 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural analysis of the G-box domain of the microcephaly protein CPAP suggests a role in centriole architecture.
Structure, 21, 2013
|
|
4M83
 
 | | Ensemble refinement of protein crystal structure (2IYF) of macrolide glycosyltransferases OleD complexed with UDP and Erythromycin A | | Descriptor: | ERYTHROMYCIN A, MAGNESIUM ION, Oleandomycin glycosyltransferase, ... | | Authors: | Wang, F, Helmich, K.E, Xu, W, Singh, S, Olmos Jr, J.L, Martinez iii, E, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2013-08-12 | | Release date: | 2013-09-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Crystal structure of macrolide glycosyltransferases OleD
To be Published
|
|
4M60
 
 | | Crystal structure of macrolide glycosyltransferases OleD | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Oleandomycin glycosyltransferase, SODIUM ION | | Authors: | Olmos Jr, J.L, Martinez III, E, Wang, F, Helmich, K.E, Singh, S, Xu, W, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2013-08-08 | | Release date: | 2013-09-04 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of macrolide glycosyltransferases OleD
To be Published
|
|
4M7P
 
 | | Ensemble refinement of protein crystal structure of macrolide glycosyltransferases OleD | | Descriptor: | Oleandomycin glycosyltransferase, SODIUM ION | | Authors: | Wang, F, Helmich, K.E, Xu, W, Singh, S, Olmos Jr, J.L, Martinez iii, E, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2013-08-12 | | Release date: | 2013-09-11 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of macrolide glycosyltransferases OleD
To be Published
|
|
4LZI
 
 | | Characterization of Solanum tuberosum Multicystatin and Significance of Core Domains | | Descriptor: | Multicystatin | | Authors: | Nissen, M.S, Kumar, G.N, Green, A.R, Knowles, N.R, Kang, C. | | Deposit date: | 2013-07-31 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Characterization of Solanum tuberosum Multicystatin and the Significance of Core Domains.
Plant Cell, 25, 2013
|
|
4N2P
 
 | | Structure of Archease from Pyrococcus horikoshii | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Desai, K.K, Bingman, C.A, Phillips Jr, G.N, Raines, R.T. | | Deposit date: | 2013-10-05 | | Release date: | 2014-01-01 | | Last modified: | 2020-09-23 | | Method: | X-RAY DIFFRACTION (1.435 Å) | | Cite: | A tRNA splicing operon: Archease endows RtcB with dual GTP/ATP cofactor specificity and accelerates RNA ligation.
Nucleic Acids Res., 42, 2014
|
|
1K8P
 
 | |
1KF1
 
 | |
4O8J
 
 | | Crystal structure of RtcA, the RNA 3'-terminal phosphate cyclase from Pyrococcus horikoshii, in complex with rACAAA3'phosphate and adenine. | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE, RNA, ... | | Authors: | Desai, K.K, Bingman, C.A, Phillips Jr, G.N, Raines, R.T. | | Deposit date: | 2013-12-27 | | Release date: | 2014-09-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure of RNA 3'-phosphate cyclase bound to substrate RNA.
Rna, 20, 2014
|
|
4O89
 
 | | Crystal structure of RtcA, the RNA 3'-terminal phosphate cyclase from Pyrococcus horikoshii. | | Descriptor: | CITRIC ACID, RNA 3'-terminal phosphate cyclase | | Authors: | Desai, K.K, Bingman, C.A, Phillips Jr, G.N, Raines, R.T. | | Deposit date: | 2013-12-26 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of RNA 3'-phosphate cyclase bound to substrate RNA.
Rna, 20, 2014
|
|
