6EUE
 
 | | Rivastigmine analogue bound to Tc ACHE. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, PENTAETHYLENE GLYCOL | | Authors: | De la Mora, E, Brazzolotto, X, Dighe, S.N, Silman, I, Weik, M, Ross, B. | | Deposit date: | 2017-10-30 | | Release date: | 2018-11-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Rivastigmine and metabolite analogues with putative Alzheimer's disease-modifying properties in a Caenorhabditis elegans model
Commun Chem, 2019
|
|
4LA3
 
 | |
6MFJ
 
 | |
8UR5
 
 | |
6M0T
 
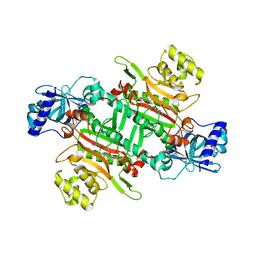 | | Crystal Structure of Lysyl-tRNA Synthetase from Plasmodium falciparum complexed with L-lysine and Cladosporin derivative (CL-2) | | Descriptor: | (3R)-3-[(R)-[(2R,6S)-6-methyloxan-2-yl]-oxidanyl-methyl]-6,8-bis(oxidanyl)-3,4-dihydroisochromen-1-one, LYSINE, Lysine--tRNA ligase | | Authors: | Babbar, P, Sharma, A, Manickam, Y. | | Deposit date: | 2020-02-22 | | Release date: | 2021-04-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Design, Synthesis, and Structural Analysis of Cladosporin-Based Inhibitors of Malaria Parasites.
Acs Infect Dis., 7, 2021
|
|
8UR7
 
 | |
7YC9
 
 | | Co-crystal structure of BTK kinase domain with inhibitor | | Descriptor: | (7~{S})-2-(4-bromanyl-3,5-dimethoxy-phenyl)-7-(1-propanoylpiperidin-4-yl)-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrimidine-3-carboxamide, 1,2-ETHANEDIOL, Tyrosine-protein kinase BTK | | Authors: | Zhou, X. | | Deposit date: | 2022-07-01 | | Release date: | 2023-05-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Discovery of BGB-8035, a Highly Selective Covalent Inhibitor of Bruton's Tyrosine Kinase for B-Cell Malignancies and Autoimmune Diseases.
J.Med.Chem., 66, 2023
|
|
7DFG
 
 | | Structure of COVID-19 RNA-dependent RNA polymerase bound to favipiravir | | Descriptor: | 6-fluoro-3-oxo-4-(5-O-phosphono-beta-D-ribofuranosyl)-3,4-dihydropyrazine-2-carboxamide, MAGNESIUM ION, Non-structural protein 7, ... | | Authors: | Li, Z, Zhou, Z, Yu, X. | | Deposit date: | 2020-11-08 | | Release date: | 2021-11-17 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis for repurpose and design of nucleotide drugs for treating COVID-19
To Be Published
|
|
7DFH
 
 | |
8BTY
 
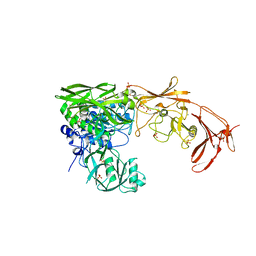 | | Structure of the active form of ScpB, the C5a-peptidase from Streptococcus agalactiae. | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, C5a peptidase, CALCIUM ION, ... | | Authors: | Kagawa, T.F, Cooney, J.C, Miclot, T, Cullen, R. | | Deposit date: | 2022-11-30 | | Release date: | 2023-11-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The 1.7 angstrom crystal structure of the C5a peptidase from Streptococcus agalactiae (ScpB) reveals an active site competent for catalysis.
Proteins, 92, 2024
|
|
7DOK
 
 | |
7DOI
 
 | |
3J8C
 
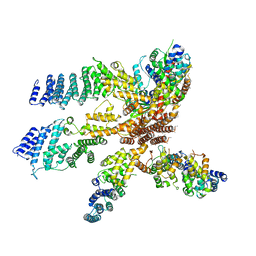 | | Model of the human eIF3 PCI-MPN octamer docked into the 43S EM map | | Descriptor: | Eukaryotic translation initiation factor 3 subunit A, Eukaryotic translation initiation factor 3 subunit C, Eukaryotic translation initiation factor 3 subunit E, ... | | Authors: | Erzberger, J.P, Ban, N. | | Deposit date: | 2014-10-08 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (11.6 Å) | | Cite: | Molecular Architecture of the 40SeIF1eIF3 Translation Initiation Complex.
Cell(Cambridge,Mass.), 158, 2014
|
|
8UK5
 
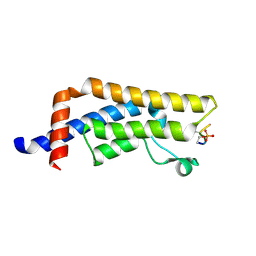 | | Crystal structure of the bromodomain of human ATAD2B in complex with histone H4S1(ph)K5ac | | Descriptor: | ATPase family AAA domain-containing protein 2B, Histone H4S1(ph)K5ac | | Authors: | Montgomery, C, Phillips, M, Nix, J.C, Glass, K.C. | | Deposit date: | 2023-10-12 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Impact of Combinatorial Histone Modifications on Acetyllysine Recognition by the ATAD2 and ATAD2B Bromodomains.
J.Med.Chem., 67, 2024
|
|
8UHL
 
 | | ATAD2B bromodomain in complex with histone H4 acetylated at lysine 12 | | Descriptor: | ATPase family AAA domain-containing protein 2B, Histone H4 | | Authors: | Phillips, M, Montgomery, C, Nix, J.C, Glass, K.C. | | Deposit date: | 2023-10-09 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Impact of Combinatorial Histone Modifications on Acetyllysine Recognition by the ATAD2 and ATAD2B Bromodomains.
J.Med.Chem., 67, 2024
|
|
5K98
 
 | | Structure of HipA-HipB-O2-O3 complex | | Descriptor: | Antitoxin HipB, DNA (5'-D(*CP*TP*AP*TP*CP*CP*CP*CP*TP*TP*AP*AP*GP*GP*GP*GP*AP*TP*AP*GP*GP*GP*A)-3'), DNA (5'-D(*TP*CP*CP*CP*TP*AP*TP*CP*CP*CP*CP*TP*TP*AP*AP*GP*GP*GP*GP*AP*TP*AP*G)-3'), ... | | Authors: | Schumacher, M. | | Deposit date: | 2016-05-31 | | Release date: | 2016-06-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.99 Å) | | Cite: | HipBA-promoter structures reveal the basis of heritable multidrug tolerance.
Nature, 524, 2015
|
|
6QP2
 
 | | Crystal structure of the PLP-bound C-S lyase from Staphylococcus hominis | | Descriptor: | Aminotransferase, PYRIDOXAL-5'-PHOSPHATE, Polyhistidine tag | | Authors: | Herman, R, Rudden, M, Wilkinson, A.J, Hanai, S, Thomas, G.H. | | Deposit date: | 2019-02-13 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The molecular basis of thioalcohol production in human body odour.
Sci Rep, 10, 2020
|
|
7CPV
 
 | | Cryo-EM structure of 80S ribosome from mouse testis | | Descriptor: | 40S ribosomal protein S10, 40S ribosomal protein S11, 40S ribosomal protein S13, ... | | Authors: | Huo, Y.G, He, X, Jiang, T, Qin, Y, Guo, X.J, Sha, J.H. | | Deposit date: | 2020-08-08 | | Release date: | 2022-02-02 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | A male germ-cell-specific ribosome controls male fertility.
Nature, 612, 2022
|
|
7CPU
 
 | | Cryo-EM structure of 80S ribosome from mouse kidney | | Descriptor: | 40S ribosomal protein S10, 40S ribosomal protein S11, 40S ribosomal protein S13, ... | | Authors: | Huo, Y.G, He, X, Jiang, T, Qin, Y, Guo, X.J, Sha, J.H. | | Deposit date: | 2020-08-08 | | Release date: | 2022-02-02 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | A male germ-cell-specific ribosome controls male fertility.
Nature, 612, 2022
|
|
7EMN
 
 | | The atomic structure of SHP2 E76A mutant | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Luo, F, Xie, J.J, Zhu, J.D, Liu, C. | | Deposit date: | 2021-04-14 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A novel partially open state of SHP2 points to a "multiple gear" regulation mechanism.
J.Biol.Chem., 296, 2021
|
|
2H8B
 
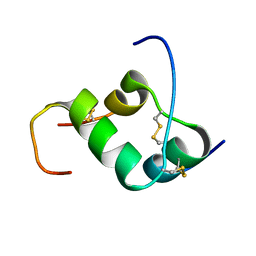 | | Solution structure of INSL3 | | Descriptor: | Insulin-like 3 | | Authors: | Rosengren, K.J, Craik, D.J, Daly, N.L. | | Deposit date: | 2006-06-07 | | Release date: | 2006-08-01 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Characterization of the LGR8 Receptor Binding Surface of Insulin-like Peptide 3
J.Biol.Chem., 281, 2006
|
|
6MFP
 
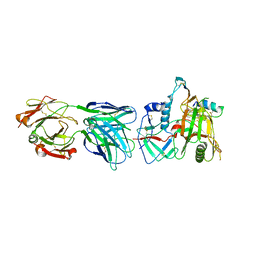 | | Crystal Structure of the RV305 C1-C2 specific ADCC potent antibody DH677.3 Fab in complex with HIV-1 clade A/E gp120 and M48U1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Tolbert, W.D, Young, B, Pazgier, M. | | Deposit date: | 2018-09-11 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Boosting with AIDSVAX B/E Enhances Env Constant Region 1 and 2 Antibody-Dependent Cellular Cytotoxicity Breadth and Potency.
J.Virol., 94, 2020
|
|
7WCY
 
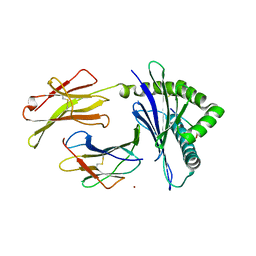 | | Crystal Structure of H-2Kb with Cryptosporidium parvum gp40/15 epitope | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, K-B alpha chain, ... | | Authors: | Wang, Y.L, Gao, M.H, Zhang, L.X, Fan, S.H. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural Analyses of a Dominant Cryptosporidium parvum Epitope Presented by H-2K b Offer New Options To Combat Cryptosporidiosis.
Mbio, 14, 2023
|
|
8V5P
 
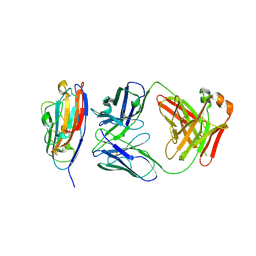 | |
8V5S
 
 | |
