7EO5
 
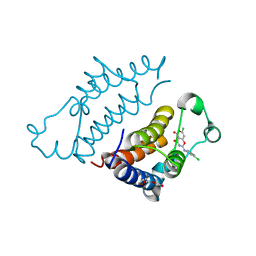 | | Crystal structure of pyrano 1,3, oxazine derivative in complex with the second bromodomain of BRD2 | | Descriptor: | 7-chloranyl-2-[(3-chlorophenyl)amino]pyrano[3,4-e][1,3]oxazine-4,5-dione, Bromodomain-containing protein 2, TRIETHYLENE GLYCOL | | Authors: | Padmanabhan, B, Arole, A, Deshmukh, P, Ashok, S. | | Deposit date: | 2021-04-21 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural investigation of a pyrano-1,3-oxazine derivative and the phenanthridinone core moiety against BRD2 bromodomains.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
6SL7
 
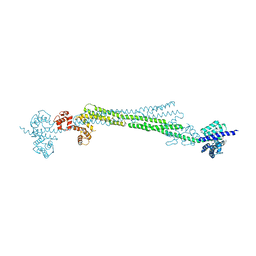 | |
6R89
 
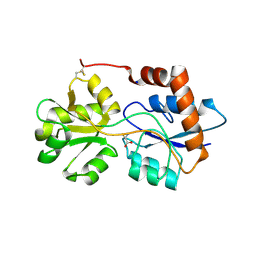 | | Structure of Arabidopsis thaliana GLR3.3 ligand-binding domain in complex with L-cysteine | | Descriptor: | CHLORIDE ION, CYSTEINE, GLYCEROL, ... | | Authors: | Alfieri, A, Pederzoli, R, Costa, A. | | Deposit date: | 2019-04-01 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structural bases for agonist diversity in anArabidopsis thalianaglutamate receptor-like channel.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6R8A
 
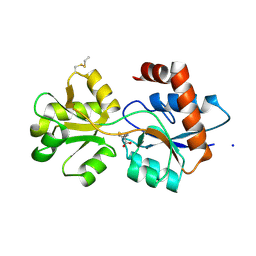 | | Structure of Arabidopsis thaliana GLR3.3 ligand-binding domain in complex with L-methionine | | Descriptor: | Glutamate receptor 3.3,Glutamate receptor 3.3, METHIONINE, SODIUM ION, ... | | Authors: | Alfieri, A, Pederzoli, R, Costa, A. | | Deposit date: | 2019-04-01 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structural bases for agonist diversity in anArabidopsis thalianaglutamate receptor-like channel.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6R88
 
 | | Structure of Arabidopsis thaliana GLR3.3 ligand-binding domain in complex with glycine | | Descriptor: | CHLORIDE ION, GLYCEROL, GLYCINE, ... | | Authors: | Alfieri, A, Pederzoli, R, Costa, A. | | Deposit date: | 2019-04-01 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structural bases for agonist diversity in anArabidopsis thalianaglutamate receptor-like channel.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6R85
 
 | | Structure of Arabidopsis thaliana GLR3.3 ligand-binding domain in complex with L-glutamate | | Descriptor: | 1,2-ETHANEDIOL, GLUTAMIC ACID, Glutamate receptor 3.3,Glutamate receptor 3.3, ... | | Authors: | Alfieri, A, Pederzoli, R, Costa, A. | | Deposit date: | 2019-03-31 | | Release date: | 2020-01-01 | | Last modified: | 2020-01-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structural bases for agonist diversity in anArabidopsis thalianaglutamate receptor-like channel.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2OQX
 
 | | Crystal Structure of the apo form of E. coli tryptophanase at 1.9 A resolution | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Goldgur, Y, Kogan, A, Gdalevsky, G, Parola, A, Cohen-Luria, R, Almog, O. | | Deposit date: | 2007-02-01 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of apo tryptophanase from Escherichia coli reveals a wide-open conformation.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
6FXN
 
 | | Crystal structure of human BAFF in complex with Fab fragment of anti-BAFF antibody belimumab | | Descriptor: | Tumor necrosis factor ligand superfamily member 13B, belimumab heavy chain, belimumab light chain | | Authors: | Lammens, A, Maskos, K, Willen, L, Jiang, X, Schneider, P. | | Deposit date: | 2018-03-09 | | Release date: | 2018-04-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A loop region of BAFF controls B cell survival and regulates recognition by different inhibitors.
Nat Commun, 9, 2018
|
|
8AYF
 
 | | Crystal structure of human Sphingosine-1-phosphate lyase 1 | | Descriptor: | ACETATE ION, GLYCEROL, Sphingosine-1-phosphate lyase 1 | | Authors: | Giardina, G, Catalano, F, Pampalone, G, Cellini, B. | | Deposit date: | 2022-09-02 | | Release date: | 2023-09-13 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Dual species sphingosine-1-phosphate lyase inhibitors to combine antifungal and anti-inflammatory activities in cystic fibrosis: a feasibility study.
Sci Rep, 13, 2023
|
|
6H1F
 
 | | Structure of the nanobody-stabilized gelsolin D187N variant (second domain) | | Descriptor: | Gelsolin, THIOCYANATE ION, gelsolin nanobody, ... | | Authors: | Hassan, A, Milani, M, Mastrangelo, E, de Rosa, M. | | Deposit date: | 2018-07-11 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Nanobody interaction unveils structure, dynamics and proteotoxicity of the Finnish-type amyloidogenic gelsolin variant.
Biochim Biophys Acta Mol Basis Dis, 1865, 2019
|
|
7P0T
 
 | |
7P0A
 
 | |
7P2B
 
 | |
8CMX
 
 | |
5NXY
 
 | | Crystal structure of OpuAC from B. subtilis in complex with Arsenobetaine | | Descriptor: | 1,2-ETHANEDIOL, 2-(trimethyl-lambda~5~-arsanyl)ethanol, Osmotically activated L-carnitine/choline ABC transporter substrate-binding protein OpuCC | | Authors: | Hofmann, T, Bremer, E, Schmitt, L, Smits, S. | | Deposit date: | 2017-05-11 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Arsenobetaine: an ecophysiologically important organoarsenical confers cytoprotection against osmotic stress and growth temperature extremes.
Environ. Microbiol., 20, 2018
|
|
6I8C
 
 | | Crystal structure of the murine beta-2-microglobulin. | | Descriptor: | Beta-2-microglobulin, TRIETHYLENE GLYCOL | | Authors: | Achour, A, Sandalova, T, Ricagno, S, Sun, R. | | Deposit date: | 2018-11-20 | | Release date: | 2019-10-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Biochemical and biophysical comparison of human and mouse beta-2 microglobulin reveals the molecular determinants of low amyloid propensity.
Febs J., 287, 2020
|
|
5NXX
 
 | | Crystal structure of OpuAC from B. subtilis in complex with Arsenobetaine | | Descriptor: | (trimethylarsonio)acetate, Glycine betaine ABC transport system glycine betaine-binding protein OpuAC | | Authors: | Hofmann, T, Bremer, E, Schmitt, L, Smits, S. | | Deposit date: | 2017-05-11 | | Release date: | 2018-03-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Arsenobetaine: an ecophysiologically important organoarsenical confers cytoprotection against osmotic stress and growth temperature extremes.
Environ. Microbiol., 20, 2018
|
|
5FDL
 
 | | Crystal Structure of K103N/Y181C Mutant HIV-1 Reverse Transcriptase (RT) in Complex with IDX899 | | Descriptor: | P51 Reverse transcriptase, P66 Reverse transcriptase, methyl (R)-(2-carbamoyl-5-chloro-1H-indol-3-yl)[3-(2-cyanoethyl)-5-methylphenyl]phosphinate | | Authors: | Dousson, C.B, Alexandre, F.-R, Convard, T, Fisher, M, Lamers, M.B.A.C, Leonard, P.M. | | Deposit date: | 2015-12-16 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Discovery of the Aryl-phospho-indole IDX899, a Highly Potent Anti-HIV Non-nucleoside Reverse Transcriptase Inhibitor.
J.Med.Chem., 59, 2016
|
|
6F6T
 
 | |
5O7G
 
 | |
5Y42
 
 | |
2GAR
 
 | | A PH-DEPENDENT STABLIZATION OF AN ACTIVE SITE LOOP OBSERVED FROM LOW AND HIGH PH CRYSTAL STRUCTURES OF MUTANT MONOMERIC GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE | | Descriptor: | GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE, PHOSPHATE ION | | Authors: | Su, Y, Yamashita, M.M, Greasley, S.E, Mullen, C.A, Shim, J.H, Jennings, P.A, Benkovic, S.J, Wilson, I.A. | | Deposit date: | 1998-05-13 | | Release date: | 1998-08-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A pH-dependent stabilization of an active site loop observed from low and high pH crystal structures of mutant monomeric glycinamide ribonucleotide transformylase at 1.8 to 1.9 A.
J.Mol.Biol., 281, 1998
|
|
7VRM
 
 | | crystal structure of BRD2-BD2 in complex with purine derivative | | Descriptor: | Bromodomain-containing protein 2, THEOPHYLLINE | | Authors: | Padmanabhan, B, Arole, A, Deshmukh, P, Ashok, S, Mathur, S. | | Deposit date: | 2021-10-23 | | Release date: | 2023-02-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural and biochemical insights into purine-based drug molecules in hBRD2 delineate a unique binding mode opening new vistas in the design of inhibitors of the BET family.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7VRO
 
 | | crystal structure of BRD2-BD1 in complex with purine derivative | | Descriptor: | Bromodomain-containing protein 2, SULFATE ION, THEOBROMINE | | Authors: | Padmanabhan, B, Arole, A, Deshmukh, P, Ashok, S, Mathur, S. | | Deposit date: | 2021-10-23 | | Release date: | 2023-02-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and biochemical insights into purine-based drug molecules in hBRD2 delineate a unique binding mode opening new vistas in the design of inhibitors of the BET family.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7VRQ
 
 | | crystal structure of BRD2-BD2 in complex with purine derivative | | Descriptor: | Bromodomain-containing protein 2, GLYCEROL, THEOBROMINE | | Authors: | Padmanabhan, B, Arole, A, Deshmukh, P, Ashok, S, Mathur, S. | | Deposit date: | 2021-10-23 | | Release date: | 2023-02-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural and biochemical insights into purine-based drug molecules in hBRD2 delineate a unique binding mode opening new vistas in the design of inhibitors of the BET family.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
