4QRR
 
 | | Crystal Structure of HLA B*3501-IPS in complex with a Delta-Beta TCR, clone 12 TCR | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-35 alpha chain, ... | | Authors: | Gras, S, Chabrol, E, Rossjohn, J. | | Deposit date: | 2014-07-02 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The molecular bases of delta / alpha beta T cell-mediated antigen recognition.
J.Exp.Med., 211, 2014
|
|
4LCW
 
 | | The structure of human MAIT TCR in complex with MR1-K43A-RL-6-Me-7OH | | Descriptor: | 1-deoxy-1-(7-hydroxy-6-methyl-2,4-dioxo-3,4-dihydropteridin-8(2H)-yl)-D-ribitol, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Patel, O, Rossjohn, J. | | Deposit date: | 2013-06-24 | | Release date: | 2013-10-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Antigen-loaded MR1 tetramers define T cell receptor heterogeneity in mucosal-associated invariant T cells.
J.Exp.Med., 210, 2013
|
|
7RSK
 
 | | The crystal structure from microfluidic crystals of glycosyl hydrolase family 2 (GH2) member from Bacteroides cellulosilyticus | | Descriptor: | Glycosyl hydrolase family 2, sugar binding domain protein | | Authors: | Kim, Y, Nocek, B, Endres, M, Joachimiak, G, Johnson, J, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2021-08-11 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure from microfluidic crystals of glycosyl hydrolase family 2 (GH2) member from Bacteroides cellulosilyticus
To Be Published
|
|
6BIN
 
 | | HLA-DRB1 in complex with Type II collagen peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HLA class II histocompatibility antigen, ... | | Authors: | Ting, Y.T, Scally, S.W, Rossjohn, J. | | Deposit date: | 2017-11-02 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.50000834 Å) | | Cite: | The interplay between citrullination and HLA-DRB1 polymorphism in shaping peptide binding hierarchies in rheumatoid arthritis.
J. Biol. Chem., 293, 2018
|
|
6BIZ
 
 | | HLA-DRB1 in complex with citrullinated Histone 2B peptide | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, HLA class II DR-beta (HLA-DR B), ... | | Authors: | Ting, Y.T, Scally, S.W, Rossjohn, J. | | Deposit date: | 2017-11-03 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The interplay between citrullination and HLA-DRB1 polymorphism in shaping peptide binding hierarchies in rheumatoid arthritis.
J. Biol. Chem., 293, 2018
|
|
2WZT
 
 | | Crystal structure of a mycobacterium aldo-keto reductase in its apo and liganded form | | Descriptor: | ALDO-KETO REDUCTASE | | Authors: | Scoble, J, McAlister, A.D, Fulton, Z, Troy, S, Byres, E, Vivian, J.P, Brammananth, R, Wilce, M.C.J, Le Nours, J, Zaker-Tabrizi, L, Coppel, R.L, Crellin, P.K, Rossjohn, J, Beddoe, T. | | Deposit date: | 2009-12-03 | | Release date: | 2010-02-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure and Comparative Functional Analyses of a Mycobacterium Aldo-Keto Reductase.
J.Mol.Biol., 398, 2010
|
|
9CI8
 
 | | T cell receptor complex | | Descriptor: | T cell receptor delta constant, T cell receptor gamma constant 1, T-cell surface glycoprotein CD3 delta chain, ... | | Authors: | Gully, B.S, Rossjohn, J. | | Deposit date: | 2024-07-02 | | Release date: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | T cell receptor complex
To Be Published
|
|
3VFW
 
 | | crystal structure of HLA B*3508 LPEP-P10Ala, peptide mutant P10-ala | | Descriptor: | Beta-2-microglobulin, LPEP peptide from EBV, P10A, ... | | Authors: | Liu, Y.C, Rossjohn, J, Gras, S. | | Deposit date: | 2012-01-10 | | Release date: | 2012-02-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Energetic Basis Underpinning T-cell Receptor Recognition of a Super-bulged Peptide Bound to a Major Histocompatibility Complex Class I Molecule.
J.Biol.Chem., 287, 2012
|
|
9CIA
 
 | | T cell receptor complex | | Descriptor: | CHOLESTEROL, T cell receptor delta constant, T cell receptor gamma constant 1, ... | | Authors: | Gully, B.S, Rossjohn, J. | | Deposit date: | 2024-07-03 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | T cell receptor complex
To Be Published
|
|
3VFO
 
 | | crystal structure of HLA B*3508 LPEP157A, HLA mutant Ala157 | | Descriptor: | Beta-2-microglobulin, LPEP peptide from EBV, LPEPLPQGQLTAY, ... | | Authors: | Liu, Y.C, Rossjohn, J, Gras, S. | | Deposit date: | 2012-01-10 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Energetic Basis Underpinning T-cell Receptor Recognition of a Super-bulged Peptide Bound to a Major Histocompatibility Complex Class I Molecule.
J.Biol.Chem., 287, 2012
|
|
3VH8
 
 | | KIR3DL1 in complex with HLA-B*5701 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Vivian, J.P, Rossjohn, J. | | Deposit date: | 2011-08-24 | | Release date: | 2011-10-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Killer cell immunoglobulin-like receptor 3DL1-mediated recognition of human leukocyte antigen B
Nature, 479, 2011
|
|
4PRN
 
 | | Crystal structure of a HLA-B*35:01-HPVG-A4 | | Descriptor: | ACETATE ION, Beta-2-microglobulin, Epstein-Barr nuclear antigen 1, ... | | Authors: | Yu Chih, L, Rossjohn, J, Gras, S. | | Deposit date: | 2014-03-06 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A Molecular Basis for the Interplay between T Cells, Viral Mutants, and Human Leukocyte Antigen Micropolymorphism.
J.Biol.Chem., 289, 2014
|
|
3VRI
 
 | | HLA-B*57:01-RVAQLENVYI in complex with abacavir | | Descriptor: | 10-mer peptide, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Vivian, J.P, Illing, P.T, McCluskey, J, Rossjohn, J. | | Deposit date: | 2012-04-11 | | Release date: | 2012-05-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Immune self-reactivity triggered by drug-modified HLA-peptide repertoire
Nature, 486, 2012
|
|
4PRD
 
 | | Crystal structure of a HLA-B*35:08-HPVG-D5 | | Descriptor: | ACETATE ION, Beta-2-microglobulin, Epstein-Barr nuclear antigen 1, ... | | Authors: | Yu Chih, L, Rossjohn, J, Gras, S. | | Deposit date: | 2014-03-05 | | Release date: | 2014-04-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A Molecular Basis for the Interplay between T Cells, Viral Mutants, and Human Leukocyte Antigen Micropolymorphism.
J.Biol.Chem., 289, 2014
|
|
3VFM
 
 | | crystal structure of HLA B*3508 LPEP155A, HLA mutant Ala155 | | Descriptor: | Beta-2-microglobulin, LPEP peptide from EBV, LPEPLPQGQLTAY, ... | | Authors: | Liu, Y.C, Rossjohn, J, Gras, S. | | Deposit date: | 2012-01-10 | | Release date: | 2012-03-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Energetic Basis Underpinning T-cell Receptor Recognition of a Super-bulged Peptide Bound to a Major Histocompatibility Complex Class I Molecule.
J.Biol.Chem., 287, 2012
|
|
3FFC
 
 | | Crystal Structure of CF34 TCR in complex with HLA-B8/FLR | | Descriptor: | Beta-2-microglobulin, CADMIUM ION, CF34 alpha chain, ... | | Authors: | Gras, S, Burrows, S.R, Kjer-Nielsen, L, Clements, C.S, Liu, Y.C, Sullivan, L.C, Brooks, A.G, Purcell, A.W, McCluskey, J, Rossjohn, J. | | Deposit date: | 2008-12-03 | | Release date: | 2009-01-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The shaping of T cell receptor recognition by self-tolerance.
Immunity, 30, 2009
|
|
6ATF
 
 | | HLA-DRB1*1402 in complex with Vimentin59-71 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scally, S.W, Ting, Y.T, Rossjohn, J. | | Deposit date: | 2017-08-29 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis for increased susceptibility of Indigenous North Americans to seropositive rheumatoid arthritis.
Ann. Rheum. Dis., 76, 2017
|
|
3VFS
 
 | | crystal structure of HLA B*3508LPEP-P5Ala , peptide mutant P5-ala | | Descriptor: | Beta-2-microglobulin, LPEP peptide from EBV, P5A, ... | | Authors: | Liu, Y.C, Rossjohn, J, Gras, S. | | Deposit date: | 2012-01-10 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Energetic Basis Underpinning T-cell Receptor Recognition of a Super-bulged Peptide Bound to a Major Histocompatibility Complex Class I Molecule.
J.Biol.Chem., 287, 2012
|
|
7T6I
 
 | | Crystal structure of HLA-DP1 in complex with pp65 peptide in reverse orientation | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCINE, ... | | Authors: | Lim, J.J, Reid, H, Rossjohn, J. | | Deposit date: | 2021-12-13 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Human T cells recognize HLA-DP-bound peptides in two orientations.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
3VFU
 
 | | crystal structure of HLA B*3508 LPEP-P7Ala, peptide mutant P7-ala | | Descriptor: | Beta-2-microglobulin, LPEP peptide from EBV, P7A, ... | | Authors: | Liu, Y.C, Rossjohn, J, Gras, S. | | Deposit date: | 2012-01-10 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Energetic Basis Underpinning T-cell Receptor Recognition of a Super-bulged Peptide Bound to a Major Histocompatibility Complex Class I Molecule.
J.Biol.Chem., 287, 2012
|
|
3VFP
 
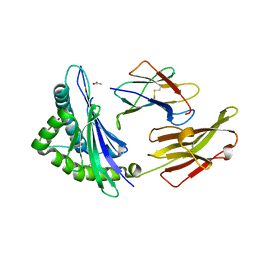 | | crystal structure of HLA B*3508 LPEP158G, HLA mutant Gly158 | | Descriptor: | ACETATE ION, Beta-2-microglobulin, LPEP peptide from EBV, ... | | Authors: | Liu, Y.C, Rossjohn, J, Gras, S. | | Deposit date: | 2012-01-10 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Energetic Basis Underpinning T-cell Receptor Recognition of a Super-bulged Peptide Bound to a Major Histocompatibility Complex Class I Molecule.
J.Biol.Chem., 287, 2012
|
|
6ATZ
 
 | | HLA-DRB1*1402 in complex with citrullinated fibrinogen peptide | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Fibrinogen beta chain, ... | | Authors: | Ting, Y.T, Scally, S.W, Rossjohn, J. | | Deposit date: | 2017-08-29 | | Release date: | 2017-09-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis for increased susceptibility of Indigenous North Americans to seropositive rheumatoid arthritis.
Ann. Rheum. Dis., 76, 2017
|
|
5J6G
 
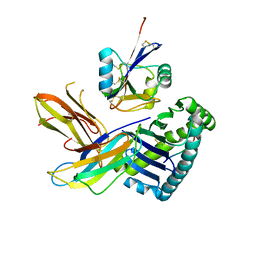 | |
6ATI
 
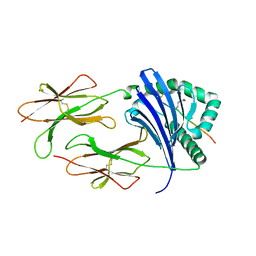 | | HLA-DRB1*1402 in complex with Vimentin-64Cit59-71 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HLA class II histocompatibility antigen, DR alpha chain, ... | | Authors: | Scally, S.W, Ting, Y.T, Rossjohn, J. | | Deposit date: | 2017-08-29 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Molecular basis for increased susceptibility of Indigenous North Americans to seropositive rheumatoid arthritis.
Ann. Rheum. Dis., 76, 2017
|
|
7U6Z
 
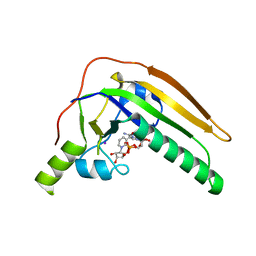 | | Pertussis toxin E129D NAD | | Descriptor: | IODIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Pertussis toxin subunit 1 | | Authors: | Littler, D.R, Beddoe, T, Pulliainen, A, Rossjohn, J. | | Deposit date: | 2022-03-06 | | Release date: | 2022-04-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.30002 Å) | | Cite: | Crystal structures of pertussis toxin with NAD + and analogs provide structural insights into the mechanism of its cytosolic ADP-ribosylation activity.
J.Biol.Chem., 298, 2022
|
|
