5F3X
 
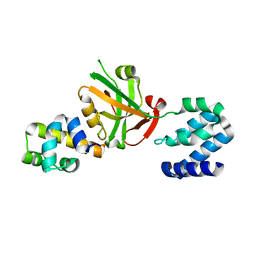 | | Crystal structure of Harmonin NPDZ1 in complex with ANKS4B SAM-PBM | | Descriptor: | Ankyrin repeat and SAM domain-containing protein 4B, CHLORIDE ION, Harmonin | | Authors: | Li, J, He, Y, Lu, Q, Zhang, M. | | Deposit date: | 2015-12-03 | | Release date: | 2016-03-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.649 Å) | | Cite: | Mechanistic Basis of Organization of the Harmonin/USH1C-Mediated Brush Border Microvilli Tip-Link Complex
Dev.Cell, 36, 2016
|
|
5YC0
 
 | | Crystal structure of LP-46/N44 | | Descriptor: | Envelope glycoprotein, LP-46 | | Authors: | Zhang, X, Wang, X, He, Y. | | Deposit date: | 2017-09-05 | | Release date: | 2018-02-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Exceptional potency and structural basis of a T1249-derived lipopeptide fusion inhibitor against HIV-1, HIV-2, and simian immunodeficiency virus
J. Biol. Chem., 293, 2018
|
|
5F3Y
 
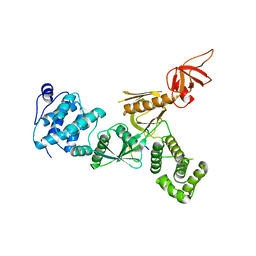 | | Crystal Structure of Myo7b N-MyTH4-FERM-SH3 in complex with Anks4b CEN | | Descriptor: | Ankyrin repeat and SAM domain-containing protein 4B, Unconventional myosin-VIIb | | Authors: | Li, J, He, Y, Lu, Q, Zhang, M. | | Deposit date: | 2015-12-03 | | Release date: | 2016-03-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.409 Å) | | Cite: | Mechanistic Basis of Organization of the Harmonin/USH1C-Mediated Brush Border Microvilli Tip-Link Complex
Dev.Cell, 36, 2016
|
|
5FUR
 
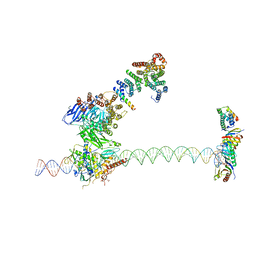 | | Structure of human TFIID-IIA bound to core promoter DNA | | Descriptor: | SUPER CORE PROMOTER, TATA-BOX-BINDING PROTEIN, TRANSCRIPTION INITIATION FACTOR IIA SUBUNIT 1, ... | | Authors: | Louder, R.K, He, Y, Lopez-Blanco, J.R, Fang, J, Chacon, P, Nogales, E. | | Deposit date: | 2016-01-29 | | Release date: | 2016-04-06 | | Last modified: | 2017-08-02 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | Structure of Promoter-Bound TFIID and Model of Human Pre-Initiation Complex Assembly.
Nature, 531, 2016
|
|
5YB4
 
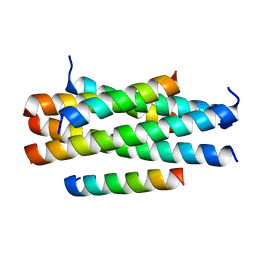 | | Crystal structure of HP23LN36KR | | Descriptor: | HP23L, N36KR | | Authors: | Zhang, X, Wang, X, He, Y. | | Deposit date: | 2017-09-03 | | Release date: | 2018-02-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insights into the Mechanisms of Action of Short-Peptide HIV-1 Fusion Inhibitors Targeting the Gp41 Pocket
Front Cell Infect Microbiol, 8, 2018
|
|
5DPD
 
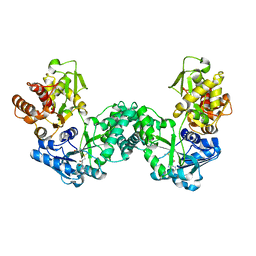 | | The structure of PKMT1 from Rickettsia prowazekii in complex with AdoMet | | Descriptor: | S-ADENOSYLMETHIONINE, protein lysine methyltransferase 1 | | Authors: | Noinaj, N, Abeykoon, A, He, Y, Yang, D.C, Buchanan, S.K. | | Deposit date: | 2015-09-12 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Insights into Substrate Recognition and Catalysis in Outer Membrane Protein B (OmpB) by Protein-lysine Methyltransferases from Rickettsia.
J.Biol.Chem., 291, 2016
|
|
1YC1
 
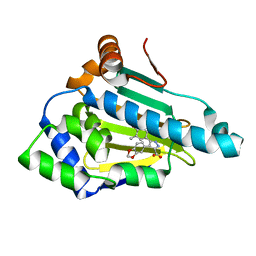 | | Crystal Structures of human HSP90alpha complexed with dihydroxyphenylpyrazoles | | Descriptor: | 4-(1,3-BENZODIOXOL-5-YL)-5-(5-ETHYL-2,4-DIHYDROXYPHENYL)-2H-PYRAZOLE-3-CARBOXYLIC ACID, Heat shock protein HSP 90-alpha | | Authors: | Kreusch, A, Han, S, Brinker, A, Zhou, V, Choi, H, He, Y, Lesley, S.A, Caldwell, J, Gu, X. | | Deposit date: | 2004-12-21 | | Release date: | 2005-02-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of human HSP90alpha-complexed with dihydroxyphenylpyrazoles.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
5DOO
 
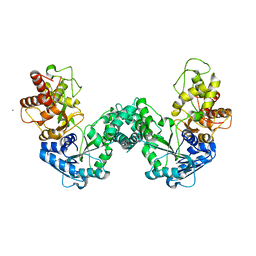 | | The structure of PKMT2 from Rickettsia typhi | | Descriptor: | CALCIUM ION, protein lysine methyltransferase 2 | | Authors: | Noinaj, N, Abeykoon, A, He, Y, Yang, D.C, Buchanan, S.K. | | Deposit date: | 2015-09-11 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.133 Å) | | Cite: | Structural Insights into Substrate Recognition and Catalysis in Outer Membrane Protein B (OmpB) by Protein-lysine Methyltransferases from Rickettsia.
J.Biol.Chem., 291, 2016
|
|
1YC4
 
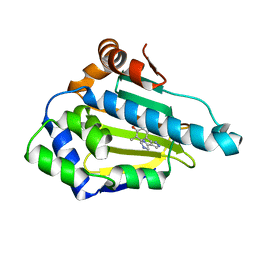 | | Crystal structure of human HSP90alpha complexed with dihydroxyphenylpyrazoles | | Descriptor: | 4-(1H-IMIDAZOL-4-YL)-3-(5-ETHYL-2,4-DIHYDROXY-PHENYL)-1H-PYRAZOLE, Heat shock protein HSP 90-alpha | | Authors: | Kreusch, A, Han, S, Brinker, A, Zhou, V, Choi, H, He, Y, Lesley, S.A, Caldwell, J, Gu, X. | | Deposit date: | 2004-12-21 | | Release date: | 2005-02-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structures of human HSP90alpha-complexed with dihydroxyphenylpyrazoles.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
1YC3
 
 | | Crystal Structure of human HSP90alpha complexed with dihydroxyphenylpyrazoles | | Descriptor: | 4-(1,3-BENZODIOXOL-5-YL)-5-(5-ETHYL-2,4-DIHYDROXYPHENYL)-2H-PYRAZOLE-3-CARBOXYLIC ACID, Heat shock protein HSP 90-alpha | | Authors: | Kreusch, A, Han, S, Brinker, A, Zhou, V, Choi, H, He, Y, Lesley, S.A, Caldwell, J, Gu, X. | | Deposit date: | 2004-12-21 | | Release date: | 2005-02-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal structures of human HSP90alpha-complexed with dihydroxyphenylpyrazoles.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
5DO0
 
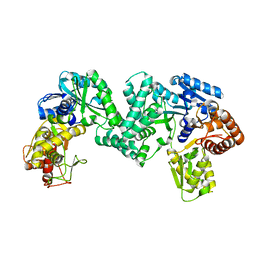 | | The structure of PKMT1 from Rickettsia prowazekii | | Descriptor: | protein lysine methyltransferase 1 | | Authors: | Noinaj, N, Abeykoon, A, He, Y, Yang, D.C, Buchanan, S.K. | | Deposit date: | 2015-09-10 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Insights into Substrate Recognition and Catalysis in Outer Membrane Protein B (OmpB) by Protein-lysine Methyltransferases from Rickettsia.
J.Biol.Chem., 291, 2016
|
|
5Y14
 
 | | Crystal structure of LP-40/N44 | | Descriptor: | LP-40, N44 | | Authors: | Zhang, X, Wang, X, He, Y. | | Deposit date: | 2017-07-19 | | Release date: | 2017-11-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.762 Å) | | Cite: | Enfuvirtide (T20)-Based Lipopeptide Is a Potent HIV-1 Cell Fusion Inhibitor: Implications for Viral Entry and Inhibition
J. Virol., 91, 2017
|
|
2OUT
 
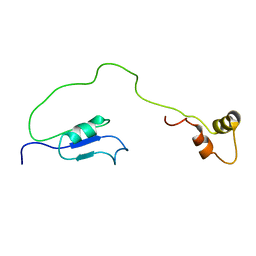 | | Solution Structure of HI1506, a Novel Two Domain Protein from Haemophilus influenzae | | Descriptor: | Mu-like prophage FluMu protein gp35, Protein HI1507 in Mu-like prophage FluMu region | | Authors: | Sari, N, He, Y, Doseeva, V, Surabian, K, Schwarz, F, Herzberg, O, Orban, J, Structure 2 Function Project (S2F) | | Deposit date: | 2007-02-12 | | Release date: | 2007-05-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of HI1506, a novel two-domain protein from Haemophilus influenzae.
Protein Sci., 16, 2007
|
|
8W6I
 
 | |
8W6J
 
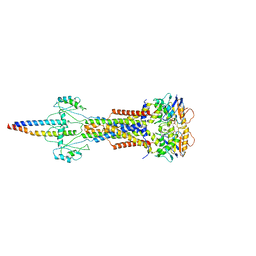 | | Cryo-EM structure of Escherichia coli Str K12 FtsE(E163Q)X/EnvC complex with ATP in peptidisc | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division ATP-binding protein FtsE, Cell division protein FtsX, ... | | Authors: | Li, J, Xu, X, He, Y, Luo, M. | | Deposit date: | 2023-08-29 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of Escherichia coli Str K12 FtsE(E163Q)X/EnvC complex with ATP in peptidisc
To Be Published
|
|
4M1Z
 
 | | Crystal structure of MycP1 with the N-terminal propeptide removed | | Descriptor: | Membrane-anchored mycosin mycp1 | | Authors: | Sun, D.M, He, Y, Wang, C.L, Zang, J.Y, Tian, C.L. | | Deposit date: | 2013-08-04 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The putative propeptide of MycP1 in mycobacterial type VII secretion system does not inhibit protease activity but improves protein stability.
Protein Cell, 4, 2013
|
|
4KB5
 
 | | Crystal structure of MycP1 from Mycobacterium smegmatis | | Descriptor: | GLYCEROL, Membrane-anchored mycosin mycp1 | | Authors: | Sun, D.M, He, Y, Tian, C.L. | | Deposit date: | 2013-04-23 | | Release date: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The putative propeptide of MycP1 in mycobacterial type VII secretion system does not inhibit protease activity but improves protein stability.
Protein Cell, 4, 2013
|
|
4NUF
 
 | | Crystal Structure of SHP/EID1 | | Descriptor: | EID1 peptide, Maltose ABC transporter periplasmic protein, Nuclear receptor subfamily 0 group B member 2 chimeric construct, ... | | Authors: | Zhi, X, Zhou, X.E, He, Y, Zechner, C, Suino-Powell, K.M, Kliewer, S.A, Melcher, K, Mangelsdorf, D.J, Xu, H.E. | | Deposit date: | 2013-12-03 | | Release date: | 2014-01-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into gene repression by the orphan nuclear receptor SHP.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6INN
 
 | |
6INV
 
 | |
6IOE
 
 | |
5XWX
 
 | |
5XX0
 
 | |
6INO
 
 | |
6INU
 
 | |
